This is a SciBERT-based model fine-tuned to perform Named Entity Recognition for drug names and adverse drug effects.
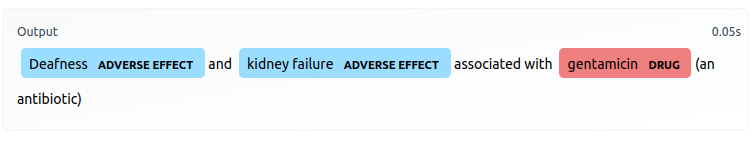
This model classifies input tokens into one of five classes:
B-DRUG: beginning of a drug entityI-DRUG: within a drug entityB-EFFECT: beginning of an AE entityI-EFFECT: within an AE entityO: outside either of the above entities
To get started using this model for inference, simply set up an NER pipeline like below:
from transformers import (AutoModelForTokenClassification,
AutoTokenizer,
pipeline,
)
model_checkpoint = "jsylee/scibert_scivocab_uncased-finetuned-ner"
model = AutoModelForTokenClassification.from_pretrained(model_checkpoint, num_labels=5,
id2label={0: 'O', 1: 'B-DRUG', 2: 'I-DRUG', 3: 'B-EFFECT', 4: 'I-EFFECT'}
)
tokenizer = AutoTokenizer.from_pretrained(model_checkpoint)
model_pipeline = pipeline(task="ner", model=model, tokenizer=tokenizer)
print( model_pipeline ("Abortion, miscarriage or uterine hemorrhage associated with misoprostol (Cytotec), a labor-inducing drug."))
SciBERT: https://huggingface.co/allenai/scibert_scivocab_uncased
- Downloads last month
- 5,561
This model does not have enough activity to be deployed to Inference API (serverless) yet. Increase its social
visibility and check back later, or deploy to Inference Endpoints (dedicated)
instead.