The viewer is disabled because this dataset repo requires arbitrary Python code execution. Please consider
removing the
loading script
and relying on
automated data support
(you can use
convert_to_parquet
from the datasets library). If this is not possible, please
open a discussion
for direct help.
M-SYNTH
M-SYNTH is a synthetic digital mammography (DM) dataset with four breast fibroglandular density distributions imaged using Monte Carlo x-ray simulations with the publicly available Virtual Imaging Clinical Trial for Regulatory Evaluation (VICTRE) toolkit.
Dataset Details
The dataset has the following characteristics:
- Breast density: dense, heterogeneously dense, scattered, fatty
- Mass radius (mm): 5.00, 7.00, 9.00
- Mass density: 1.0, 1.06, 1.1 (ratio of radiodensity of the mass to that of fibroglandular tissue)
- Relative dose: 20%, 40%, 60%, 80%, 100% of the clinically recommended dose for each density
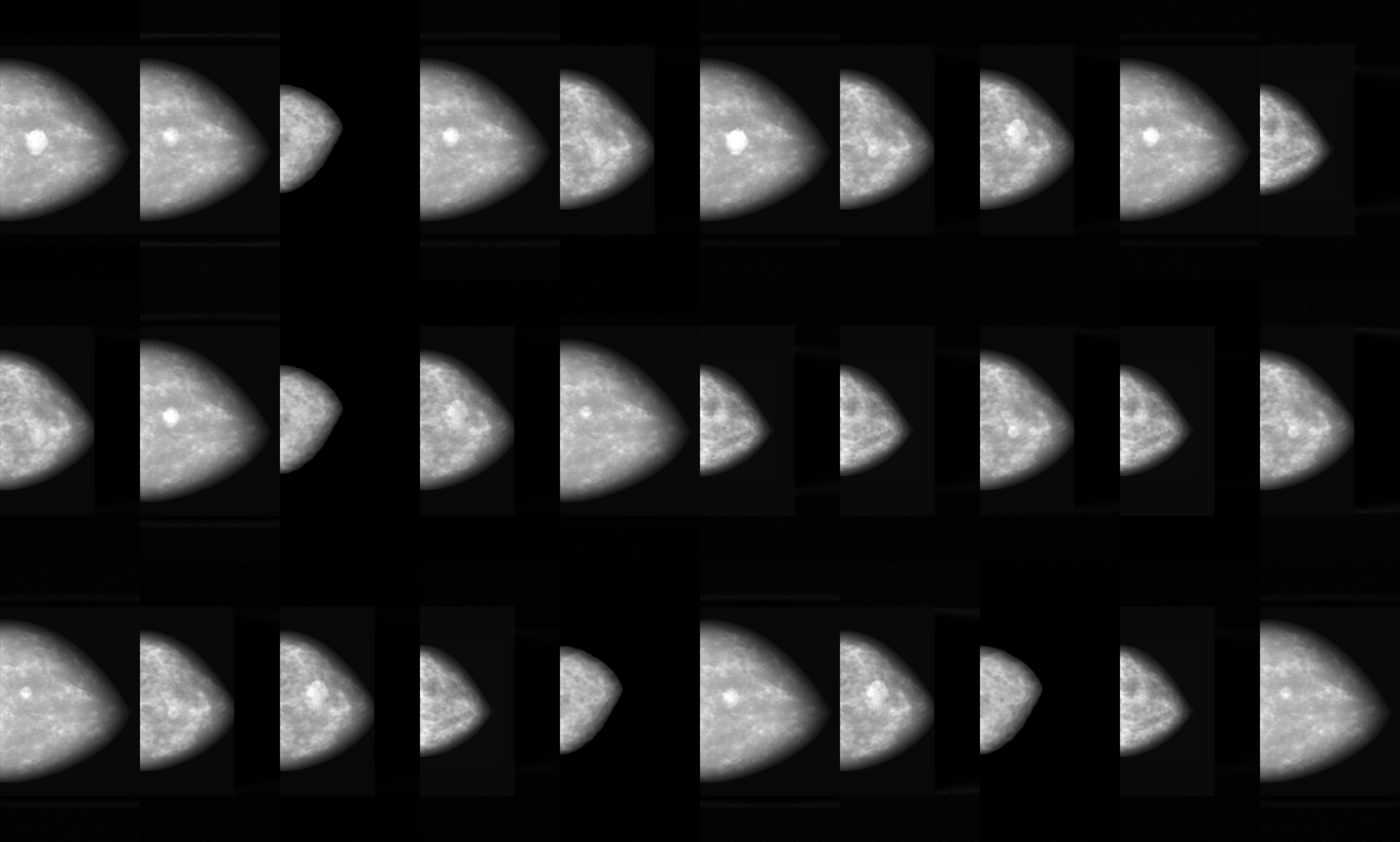
Dataset Description
- Curated by: Elena Sizikova, Niloufar Saharkhiz, Diksha Sharma, Miguel Lago, Berkman Sahiner, Jana Gut Delfino, Aldo Badano
- License: Creative Commons 1.0 Universal License (CC0)
Dataset Sources
- Code: https://github.com/DIDSR/msynth-release
- Paper: https://arxiv.org/pdf/2310.18494.pdf
- Demo: https://github.com/DIDSR/msynth-release/tree/master/examples
Uses
M-SYNTH is intended to facilitate testing of AI with pre-computed synthetic mammography data.
Direct Use
M-SYNTH can be used to evaluate the effect of mass size and density, breast density, and dose on AI performance in lesion detection. M-SYNTH can be used to either train or test pre-trained AI models.
Out-of-Scope Use
M-SYNTH cannot be used in lieu of real patient examples to make performance determinations.
Dataset Structure
M-SYNTH is organized into a directory structure that indicates the parameters. The folder
device_data_VICTREPhantoms_spic_[LESION_DENSITY]/[DOSE]/[BREAST_DENSITY]/2/[LESION_SIZE]/SIM/P2_[LESION_SIZE]_[BREAST_DENSITY].8337609.[PHANTOM_FILE_ID]/[PHANTOM_FILEID]/
contains image files imaged with the specified parameters. Note that only examples with odd PHANTOM_FILEID contain lesions, others do not.
$ tree data/device_data_VICTREPhantoms_spic_1.0/1.02e10/hetero/2/5.0/SIM/P2_5.0_hetero.8337609.1/1/
data/device_data_VICTREPhantoms_spic_1.0/1.02e10/hetero/2/5.0/SIM/P2_5.0_hetero.8337609.1/1/
├── DICOM_dm
│ └── 000.dcm
├── projection_DM1.loc
├── projection_DM1.mhd
└── projection_DM1.raw
Each folder contains mammogram data that can be read from .raw format (.mhd contains supporting data), or DICOM (.dcm) format. Coordinates of lesions can be found in .loc files. Segmentations are stored in .raw format and can be found in data/segmentation_masks/* .
See Github for examples of how to access the files, and examples for code to load each type of file.
Bias, Risks, and Limitations
Simulation-based testing is constrained to the parameter variability represented in the object model and the acquisition system. There is a risk of misjudging model performance if the simulated examples do not capture the variability in real patients. Please see the paper for a full discussion of biases, risks, and limitations.
How to use it
The msynth dataset is a very large dataset so for most use cases it is recommended to make use of the streaming API of datasets.
The msynth dataset has three configurations: 1) device_data, 2) segmentation_mask, and 3) metadata
You can load and iterate through the dataset using the configurations with the following lines of code:
from datasets import load_dataset
ds = load_dataset("didsr/msynth", 'device_data') # For device data for all breast density, mass redius, mass density, and relative dose, change configuration to 'segmentation_mask' and 'metadata' to load the segmentation masks and bound information
print(ds_data["device_data"])
# A sample data instance
{'Raw': '~\\.cache\\huggingface\\datasets\\downloads\\extracted\\59384cf05fc44e8c0cb23bb19e1fcd8f0c39720b282109d204a85561fe66bdb1\\SIM\\P2_5.0_fatty.8336179.1\\1\\projection_DM1.raw',
'mhd': '~/.cache/huggingface/datasets/downloads/extracted/59384cf05fc44e8c0cb23bb19e1fcd8f0c39720b282109d204a85561fe66bdb1/SIM/P2_5.0_fatty.8336179.1/1\\projection_DM1.mhd',
'loc': '~/.cache/huggingface/datasets/downloads/extracted/59384cf05fc44e8c0cb23bb19e1fcd8f0c39720b282109d204a85561fe66bdb1/SIM/P2_5.0_fatty.8336179.1/1\\projection_DM1.loc',
'dcm': '~/.cache/huggingface/datasets/downloads/extracted/59384cf05fc44e8c0cb23bb19e1fcd8f0c39720b282109d204a85561fe66bdb1/SIM/P2_5.0_fatty.8336179.1/1\\DICOM_dm\\000.dcm',
'density': 'fatty',
'mass_radius': 5.0}
Msynth dataset can also be loaded using custom breast density, mass redius, mass density, and relative dose information
from datasets import load_dataset
# Dataset properties. change to 'all' to include all the values of breast density, mass redius, mass density, and relative dose information
config_kwargs = {
"lesion_density": ["1.0"],
"dose": ["20%"],
"density": ["fatty"],
"size": ["5.0"]
}
# Loading device data
ds_data = load_dataset("didsr/msynth", 'device_data', **config_kwargs)
# Loading segmentation-mask
ds_seg = load_dataset("didsr/msynth", 'segmentation_mask', **config_kwargs)
The meta data can also be loaded using the datasets API. An example of using metadata is given in Demo: https://github.com/DIDSR/msynth-release/tree/master/examples
from datasets import load_dataset
# Loading metadata
ds_meta = load_dataset("didsr/msynth", 'metadata')
# A sample data instance
ds_meta['metadata'][0]
# Output
{'fatty': '~\\.cache\\huggingface\\datasets\\downloads\\extracted\\3ea85fc6b3fcc253ac8550b5d1b21db406ca9a59ea125ff8fc63d9b754c88348\\bounds\\bounds_fatty.npy',
'dense': '~\\.cache\\huggingface\\datasets\\downloads\\extracted\\3ea85fc6b3fcc253ac8550b5d1b21db406ca9a59ea125ff8fc63d9b754c88348\\bounds\\bounds_dense.npy',
'hetero': '~\\.cache\\huggingface\\datasets\\downloads\\extracted\\3ea85fc6b3fcc253ac8550b5d1b21db406ca9a59ea125ff8fc63d9b754c88348\\bounds\\bounds_hetero.npy',
'scattered': '~\\.cache\\huggingface\\datasets\\downloads\\extracted\\3ea85fc6b3fcc253ac8550b5d1b21db406ca9a59ea125ff8fc63d9b754c88348\\bounds\\bounds_scattered.npy'}
Citation
@article{sizikova2023knowledge,
title={Knowledge-based in silico models and dataset for the comparative evaluation of mammography AI for a range of breast characteristics, lesion conspicuities and doses},
author={Sizikova, Elena and Saharkhiz, Niloufar and Sharma, Diksha and Lago, Miguel and Sahiner, Berkman and Delfino, Jana G. and Badano, Aldo},
journal={Advances in Neural Information Processing Systems},
volume={},
pages={},
year={2023}
}
Related Links
- Virtual Imaging Clinical Trial for Regulatory Evaluation (VICTRE).
- FDA Catalog of Regulatory Science Tools to Help Assess New Medical Devices.
- A. Badano, C. G. Graff, A. Badal, D. Sharma, R. Zeng, F. W. Samuelson, S. Glick, K. J. Myers. Evaluation of Digital Breast Tomosynthesis as Replacement of Full-Field Digital Mammography Using an In Silico Imaging Trial. JAMA Network Open 2018.
- A. Badano, M. Lago, E. Sizikova, J. G. Delfino, S. Guan, M. A. Anastasio, B. Sahiner. The stochastic digital human is now enrolling for in silico imaging trials—methods and tools for generating digital cohorts. Progress in Biomedical Engineering 2023.
- E. Sizikova, N. Saharkhiz, D. Sharma, M. Lago, B. Sahiner, J. G. Delfino, A. Badano. Knowledge-based in silico models and dataset for the comparative evaluation of mammography AI. NeurIPS 2023 Workshop on Synthetic Data Generation with Generative AI.
- Downloads last month
- 220