hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
d0000eb66b25d89c2d8a5d46ce7f89d88ad58f91 | 14,725 | ipynb | Jupyter Notebook | Lectures/09_StrainGage.ipynb | eiriniflorou/GWU-MAE3120_2022 | 52cd589c4cfcb0dda357c326cc60c2951cedca3b | [
"BSD-3-Clause"
] | 5 | 2022-01-11T17:38:12.000Z | 2022-02-05T05:02:50.000Z | Lectures/09_StrainGage.ipynb | eiriniflorou/GWU-MAE3120_2022 | 52cd589c4cfcb0dda357c326cc60c2951cedca3b | [
"BSD-3-Clause"
] | null | null | null | Lectures/09_StrainGage.ipynb | eiriniflorou/GWU-MAE3120_2022 | 52cd589c4cfcb0dda357c326cc60c2951cedca3b | [
"BSD-3-Clause"
] | 9 | 2022-01-13T17:55:14.000Z | 2022-03-24T14:41:03.000Z | 38.955026 | 518 | 0.584652 | [
[
[
"# 09 Strain Gage\n\nThis is one of the most commonly used sensor. It is used in many transducers. Its fundamental operating principle is fairly easy to understand and it will be the purpose of this lecture. \n\nA strain gage is essentially a thin wire that is wrapped on film of plastic. \n<img src=\"img/StrainGage.png\" width=\"200\">\nThe strain gage is then mounted (glued) on the part for which the strain must be measured. \n<img src=\"img/Strain_gauge_2.jpg\" width=\"200\">\n\n## Stress, Strain\nWhen a beam is under axial load, the axial stress, $\\sigma_a$, is defined as:\n\\begin{align*}\n\\sigma_a = \\frac{F}{A}\n\\end{align*}\nwith $F$ the axial load, and $A$ the cross sectional area of the beam under axial load.\n\n<img src=\"img/BeamUnderStrain.png\" width=\"200\">\n\nUnder the load, the beam of length $L$ will extend by $dL$, giving rise to the definition of strain, $\\epsilon_a$:\n\\begin{align*}\n\\epsilon_a = \\frac{dL}{L}\n\\end{align*}\nThe beam will also contract laterally: the cross sectional area is reduced by $dA$. This results in a transverval strain $\\epsilon_t$. The transversal and axial strains are related by the Poisson's ratio:\n\\begin{align*}\n\\nu = - \\frac{\\epsilon_t }{\\epsilon_a}\n\\end{align*}\nFor a metal the Poission's ratio is typically $\\nu = 0.3$, for an incompressible material, such as rubber (or water), $\\nu = 0.5$.\n\nWithin the elastic limit, the axial stress and axial strain are related through Hooke's law by the Young's modulus, $E$:\n\\begin{align*}\n\\sigma_a = E \\epsilon_a\n\\end{align*}\n\n<img src=\"img/ElasticRegime.png\" width=\"200\">",
"_____no_output_____"
],
[
"## Resistance of a wire\n\nThe electrical resistance of a wire $R$ is related to its physical properties (the electrical resistiviy, $\\rho$ in $\\Omega$/m) and its geometry: length $L$ and cross sectional area $A$.\n\n\\begin{align*}\nR = \\frac{\\rho L}{A}\n\\end{align*}\n\nMathematically, the change in wire dimension will result inchange in its electrical resistance. This can be derived from first principle:\n\\begin{align}\n\\frac{dR}{R} = \\frac{d\\rho}{\\rho} + \\frac{dL}{L} - \\frac{dA}{A}\n\\end{align}\nIf the wire has a square cross section, then:\n\\begin{align*}\nA & = L'^2 \\\\\n\\frac{dA}{A} & = \\frac{d(L'^2)}{L'^2} = \\frac{2L'dL'}{L'^2} = 2 \\frac{dL'}{L'}\n\\end{align*}\nWe have related the change in cross sectional area to the transversal strain.\n\\begin{align*}\n\\epsilon_t = \\frac{dL'}{L'}\n\\end{align*}\nUsing the Poisson's ratio, we can relate then relate the change in cross-sectional area ($dA/A$) to axial strain $\\epsilon_a = dL/L$.\n\\begin{align*}\n\\epsilon_t &= - \\nu \\epsilon_a \\\\\n\\frac{dL'}{L'} &= - \\nu \\frac{dL}{L} \\; \\text{or}\\\\\n\\frac{dA}{A} & = 2\\frac{dL'}{L'} = -2 \\nu \\frac{dL}{L}\n\\end{align*}\nFinally we can substitute express $dA/A$ in eq. for $dR/R$ and relate change in resistance to change of wire geometry, remembering that for a metal $\\nu =0.3$:\n\\begin{align}\n\\frac{dR}{R} & = \\frac{d\\rho}{\\rho} + \\frac{dL}{L} - \\frac{dA}{A} \\\\\n& = \\frac{d\\rho}{\\rho} + \\frac{dL}{L} - (-2\\nu \\frac{dL}{L}) \\\\\n& = \\frac{d\\rho}{\\rho} + 1.6 \\frac{dL}{L} = \\frac{d\\rho}{\\rho} + 1.6 \\epsilon_a\n\\end{align}\nIt also happens that for most metals, the resistivity increases with axial strain. In general, one can then related the change in resistance to axial strain by defining the strain gage factor:\n\\begin{align}\nS = 1.6 + \\frac{d\\rho}{\\rho}\\cdot \\frac{1}{\\epsilon_a}\n\\end{align}\nand finally, we have:\n\\begin{align*}\n\\frac{dR}{R} = S \\epsilon_a\n\\end{align*}\n$S$ is materials dependent and is typically equal to 2.0 for most commercially availabe strain gages. It is dimensionless.\n\nStrain gages are made of thin wire that is wraped in several loops, effectively increasing the length of the wire and therefore the sensitivity of the sensor.\n\n_Question:\n\nExplain why a longer wire is necessary to increase the sensitivity of the sensor_.\n\nMost commercially available strain gages have a nominal resistance (resistance under no load, $R_{ini}$) of 120 or 350 $\\Omega$.\n\nWithin the elastic regime, strain is typically within the range $10^{-6} - 10^{-3}$, in fact strain is expressed in unit of microstrain, with a 1 microstrain = $10^{-6}$. Therefore, changes in resistances will be of the same order. If one were to measure resistances, we will need a dynamic range of 120 dB, whih is typically very expensive. Instead, one uses the Wheatstone bridge to transform the change in resistance to a voltage, which is easier to measure and does not require such a large dynamic range.",
"_____no_output_____"
],
[
"## Wheatstone bridge:\n<img src=\"img/WheatstoneBridge.png\" width=\"200\">\n\nThe output voltage is related to the difference in resistances in the bridge:\n\\begin{align*}\n\\frac{V_o}{V_s} = \\frac{R_1R_3-R_2R_4}{(R_1+R_4)(R_2+R_3)}\n\\end{align*}\n\nIf the bridge is balanced, then $V_o = 0$, it implies: $R_1/R_2 = R_4/R_3$.\n\nIn practice, finding a set of resistors that balances the bridge is challenging, and a potentiometer is used as one of the resistances to do minor adjustement to balance the bridge. If one did not do the adjustement (ie if we did not zero the bridge) then all the measurement will have an offset or bias that could be removed in a post-processing phase, as long as the bias stayed constant.\n\nIf each resistance $R_i$ is made to vary slightly around its initial value, ie $R_i = R_{i,ini} + dR_i$. For simplicity, we will assume that the initial value of the four resistances are equal, ie $R_{1,ini} = R_{2,ini} = R_{3,ini} = R_{4,ini} = R_{ini}$. This implies that the bridge was initially balanced, then the output voltage would be:\n\n\\begin{align*}\n\\frac{V_o}{V_s} = \\frac{1}{4} \\left( \\frac{dR_1}{R_{ini}} - \\frac{dR_2}{R_{ini}} + \\frac{dR_3}{R_{ini}} - \\frac{dR_4}{R_{ini}} \\right)\n\\end{align*}\n\nNote here that the changes in $R_1$ and $R_3$ have a positive effect on $V_o$, while the changes in $R_2$ and $R_4$ have a negative effect on $V_o$. In practice, this means that is a beam is a in tension, then a strain gage mounted on the branch 1 or 3 of the Wheatstone bridge will produce a positive voltage, while a strain gage mounted on branch 2 or 4 will produce a negative voltage. One takes advantage of this to increase sensitivity to measure strain.\n\n### Quarter bridge\nOne uses only one quarter of the bridge, ie strain gages are only mounted on one branch of the bridge.\n\n\\begin{align*}\n\\frac{V_o}{V_s} = \\pm \\frac{1}{4} \\epsilon_a S\n\\end{align*}\nSensitivity, $G$:\n\\begin{align*}\nG = \\frac{V_o}{\\epsilon_a} = \\pm \\frac{1}{4}S V_s\n\\end{align*}\n\n\n### Half bridge\nOne uses half of the bridge, ie strain gages are mounted on two branches of the bridge.\n\n\\begin{align*}\n\\frac{V_o}{V_s} = \\pm \\frac{1}{2} \\epsilon_a S\n\\end{align*}\n\n### Full bridge\n\nOne uses of the branches of the bridge, ie strain gages are mounted on each branch.\n\n\\begin{align*}\n\\frac{V_o}{V_s} = \\pm \\epsilon_a S\n\\end{align*}\n\nTherefore, as we increase the order of bridge, the sensitivity of the instrument increases. However, one should be carefull how we mount the strain gages as to not cancel out their measurement.",
"_____no_output_____"
],
[
"_Exercise_\n\n1- Wheatstone bridge\n\n<img src=\"img/WheatstoneBridge.png\" width=\"200\">\n\n> How important is it to know \\& match the resistances of the resistors you employ to create your bridge?\n> How would you do that practically?\n> Assume $R_1=120\\,\\Omega$, $R_2=120\\,\\Omega$, $R_3=120\\,\\Omega$, $R_4=110\\,\\Omega$, $V_s=5.00\\,\\text{V}$. What is $V_\\circ$?",
"_____no_output_____"
]
],
[
[
"Vs = 5.00\nVo = (120**2-120*110)/(230*240) * Vs\nprint('Vo = ',Vo, ' V')",
"Vo = 0.10869565217391304 V\n"
],
[
"# typical range in strain a strain gauge can measure\n# 1 -1000 micro-Strain\nAxialStrain = 1000*10**(-6) # axial strain\nStrainGageFactor = 2\nR_ini = 120 # Ohm\nR_1 = R_ini+R_ini*StrainGageFactor*AxialStrain\nprint(R_1)\nVo = (120**2-120*(R_1))/((120+R_1)*240) * Vs\nprint('Vo = ', Vo, ' V')",
"120.24\nVo = -0.002497502497502434 V\n"
]
],
[
[
"> How important is it to know \\& match the resistances of the resistors you employ to create your bridge?\n> How would you do that practically?\n> Assume $R_1= R_2 =R_3=120\\,\\Omega$, $R_4=120.01\\,\\Omega$, $V_s=5.00\\,\\text{V}$. What is $V_\\circ$?",
"_____no_output_____"
]
],
[
[
"Vs = 5.00\nVo = (120**2-120*120.01)/(240.01*240) * Vs\nprint(Vo)",
"-0.00010416232656978944\n"
]
],
[
[
"2- Strain gage 1:\n\nOne measures the strain on a bridge steel beam. The modulus of elasticity is $E=190$ GPa. Only one strain gage is mounted on the bottom of the beam; the strain gage factor is $S=2.02$.\n\n> a) What kind of electronic circuit will you use? Draw a sketch of it.\n\n> b) Assume all your resistors including the unloaded strain gage are balanced and measure $120\\,\\Omega$, and that the strain gage is at location $R_2$. The supply voltage is $5.00\\,\\text{VDC}$. Will $V_\\circ$ be positive or negative when a downward load is added?",
"_____no_output_____"
],
[
"In practice, we cannot have all resistances = 120 $\\Omega$. at zero load, the bridge will be unbalanced (show $V_o \\neq 0$). How could we balance our bridge?\n\nUse a potentiometer to balance bridge, for the load cell, we ''zero'' the instrument.\n\nOther option to zero-out our instrument? Take data at zero-load, record the voltage, $V_{o,noload}$. Substract $V_{o,noload}$ to my data.",
"_____no_output_____"
],
[
"> c) For a loading in which $V_\\circ = -1.25\\,\\text{mV}$, calculate the strain $\\epsilon_a$ in units of microstrain.",
"_____no_output_____"
],
[
"\\begin{align*}\n\\frac{V_o}{V_s} & = - \\frac{1}{4} \\epsilon_a S\\\\\n\\epsilon_a & = -\\frac{4}{S} \\frac{V_o}{V_s}\n\\end{align*}",
"_____no_output_____"
]
],
[
[
"S = 2.02\nVo = -0.00125\nVs = 5\neps_a = -1*(4/S)*(Vo/Vs)\nprint(eps_a)",
"0.0004950495049504951\n"
]
],
[
[
"> d) Calculate the axial stress (in MPa) in the beam under this load.",
"_____no_output_____"
],
[
"> e) You now want more sensitivity in your measurement, you install a second strain gage on to",
"_____no_output_____"
],
[
"p of the beam. Which resistor should you use for this second active strain gage?\n\n> f) With this new setup and the same applied load than previously, what should be the output voltage?",
"_____no_output_____"
],
[
"3- Strain Gage with Long Lead Wires \n\n<img src=\"img/StrainGageLongWires.png\" width=\"360\">\n\nA quarter bridge strain gage Wheatstone bridge circuit is constructed with $120\\,\\Omega$ resistors and a $120\\,\\Omega$ strain gage. For this practical application, the strain gage is located very far away form the DAQ station and the lead wires to the strain gage are $10\\,\\text{m}$ long and the lead wire have a resistance of $0.080\\,\\Omega/\\text{m}$. The lead wire resistance can lead to problems since $R_{lead}$ changes with temperature.\n\n> Design a modified circuit that will cancel out the effect of the lead wires.",
"_____no_output_____"
],
[
"## Homework\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0002eb938681f1aa86606ced02f1a76ee95018f | 10,708 | ipynb | Jupyter Notebook | nbs/43_tabular.learner.ipynb | NickVlasov/fastai | 2daa6658b467e795bdef16c980aa7ddfbe55d09c | [
"Apache-2.0"
] | 5 | 2020-08-27T00:52:27.000Z | 2022-03-31T02:46:05.000Z | nbs/43_tabular.learner.ipynb | NickVlasov/fastai | 2daa6658b467e795bdef16c980aa7ddfbe55d09c | [
"Apache-2.0"
] | null | null | null | nbs/43_tabular.learner.ipynb | NickVlasov/fastai | 2daa6658b467e795bdef16c980aa7ddfbe55d09c | [
"Apache-2.0"
] | 2 | 2021-04-17T03:33:21.000Z | 2022-02-25T19:32:34.000Z | 33.254658 | 416 | 0.593108 | [
[
[
"#export\nfrom fastai.basics import *\nfrom fastai.tabular.core import *\nfrom fastai.tabular.model import *",
"_____no_output_____"
],
[
"from fastai.tabular.data import *",
"_____no_output_____"
],
[
"#hide\nfrom nbdev.showdoc import *",
"_____no_output_____"
],
[
"#default_exp tabular.learner",
"_____no_output_____"
]
],
[
[
"# Tabular learner\n\n> The function to immediately get a `Learner` ready to train for tabular data",
"_____no_output_____"
],
[
"The main function you probably want to use in this module is `tabular_learner`. It will automatically create a `TabulaModel` suitable for your data and infer the irght loss function. See the [tabular tutorial](http://docs.fast.ai/tutorial.tabular) for an example of use in context.",
"_____no_output_____"
],
[
"## Main functions",
"_____no_output_____"
]
],
[
[
"#export\n@log_args(but_as=Learner.__init__)\nclass TabularLearner(Learner):\n \"`Learner` for tabular data\"\n def predict(self, row):\n tst_to = self.dls.valid_ds.new(pd.DataFrame(row).T)\n tst_to.process()\n tst_to.conts = tst_to.conts.astype(np.float32)\n dl = self.dls.valid.new(tst_to)\n inp,preds,_,dec_preds = self.get_preds(dl=dl, with_input=True, with_decoded=True)\n i = getattr(self.dls, 'n_inp', -1)\n b = (*tuplify(inp),*tuplify(dec_preds))\n full_dec = self.dls.decode((*tuplify(inp),*tuplify(dec_preds)))\n return full_dec,dec_preds[0],preds[0]",
"_____no_output_____"
],
[
"show_doc(TabularLearner, title_level=3)",
"_____no_output_____"
]
],
[
[
"It works exactly as a normal `Learner`, the only difference is that it implements a `predict` method specific to work on a row of data.",
"_____no_output_____"
]
],
[
[
"#export\n@log_args(to_return=True, but_as=Learner.__init__)\n@delegates(Learner.__init__)\ndef tabular_learner(dls, layers=None, emb_szs=None, config=None, n_out=None, y_range=None, **kwargs):\n \"Get a `Learner` using `dls`, with `metrics`, including a `TabularModel` created using the remaining params.\"\n if config is None: config = tabular_config()\n if layers is None: layers = [200,100]\n to = dls.train_ds\n emb_szs = get_emb_sz(dls.train_ds, {} if emb_szs is None else emb_szs)\n if n_out is None: n_out = get_c(dls)\n assert n_out, \"`n_out` is not defined, and could not be infered from data, set `dls.c` or pass `n_out`\"\n if y_range is None and 'y_range' in config: y_range = config.pop('y_range')\n model = TabularModel(emb_szs, len(dls.cont_names), n_out, layers, y_range=y_range, **config)\n return TabularLearner(dls, model, **kwargs)",
"_____no_output_____"
]
],
[
[
"If your data was built with fastai, you probably won't need to pass anything to `emb_szs` unless you want to change the default of the library (produced by `get_emb_sz`), same for `n_out` which should be automatically inferred. `layers` will default to `[200,100]` and is passed to `TabularModel` along with the `config`.\n\nUse `tabular_config` to create a `config` and cusotmize the model used. There is just easy access to `y_range` because this argument is often used.\n\nAll the other arguments are passed to `Learner`.",
"_____no_output_____"
]
],
[
[
"path = untar_data(URLs.ADULT_SAMPLE)\ndf = pd.read_csv(path/'adult.csv')\ncat_names = ['workclass', 'education', 'marital-status', 'occupation', 'relationship', 'race']\ncont_names = ['age', 'fnlwgt', 'education-num']\nprocs = [Categorify, FillMissing, Normalize]\ndls = TabularDataLoaders.from_df(df, path, procs=procs, cat_names=cat_names, cont_names=cont_names, \n y_names=\"salary\", valid_idx=list(range(800,1000)), bs=64)\nlearn = tabular_learner(dls)",
"_____no_output_____"
],
[
"#hide\ntst = learn.predict(df.iloc[0])",
"_____no_output_____"
],
[
"#hide\n#test y_range is passed\nlearn = tabular_learner(dls, y_range=(0,32))\nassert isinstance(learn.model.layers[-1], SigmoidRange)\ntest_eq(learn.model.layers[-1].low, 0)\ntest_eq(learn.model.layers[-1].high, 32)\n\nlearn = tabular_learner(dls, config = tabular_config(y_range=(0,32)))\nassert isinstance(learn.model.layers[-1], SigmoidRange)\ntest_eq(learn.model.layers[-1].low, 0)\ntest_eq(learn.model.layers[-1].high, 32)",
"_____no_output_____"
],
[
"#export\n@typedispatch\ndef show_results(x:Tabular, y:Tabular, samples, outs, ctxs=None, max_n=10, **kwargs):\n df = x.all_cols[:max_n]\n for n in x.y_names: df[n+'_pred'] = y[n][:max_n].values\n display_df(df)",
"_____no_output_____"
]
],
[
[
"## Export -",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.export import notebook2script\nnotebook2script()",
"Converted 00_torch_core.ipynb.\nConverted 01_layers.ipynb.\nConverted 02_data.load.ipynb.\nConverted 03_data.core.ipynb.\nConverted 04_data.external.ipynb.\nConverted 05_data.transforms.ipynb.\nConverted 06_data.block.ipynb.\nConverted 07_vision.core.ipynb.\nConverted 08_vision.data.ipynb.\nConverted 09_vision.augment.ipynb.\nConverted 09b_vision.utils.ipynb.\nConverted 09c_vision.widgets.ipynb.\nConverted 10_tutorial.pets.ipynb.\nConverted 11_vision.models.xresnet.ipynb.\nConverted 12_optimizer.ipynb.\nConverted 13_callback.core.ipynb.\nConverted 13a_learner.ipynb.\nConverted 13b_metrics.ipynb.\nConverted 14_callback.schedule.ipynb.\nConverted 14a_callback.data.ipynb.\nConverted 15_callback.hook.ipynb.\nConverted 15a_vision.models.unet.ipynb.\nConverted 16_callback.progress.ipynb.\nConverted 17_callback.tracker.ipynb.\nConverted 18_callback.fp16.ipynb.\nConverted 18a_callback.training.ipynb.\nConverted 19_callback.mixup.ipynb.\nConverted 20_interpret.ipynb.\nConverted 20a_distributed.ipynb.\nConverted 21_vision.learner.ipynb.\nConverted 22_tutorial.imagenette.ipynb.\nConverted 23_tutorial.vision.ipynb.\nConverted 24_tutorial.siamese.ipynb.\nConverted 24_vision.gan.ipynb.\nConverted 30_text.core.ipynb.\nConverted 31_text.data.ipynb.\nConverted 32_text.models.awdlstm.ipynb.\nConverted 33_text.models.core.ipynb.\nConverted 34_callback.rnn.ipynb.\nConverted 35_tutorial.wikitext.ipynb.\nConverted 36_text.models.qrnn.ipynb.\nConverted 37_text.learner.ipynb.\nConverted 38_tutorial.text.ipynb.\nConverted 40_tabular.core.ipynb.\nConverted 41_tabular.data.ipynb.\nConverted 42_tabular.model.ipynb.\nConverted 43_tabular.learner.ipynb.\nConverted 44_tutorial.tabular.ipynb.\nConverted 45_collab.ipynb.\nConverted 46_tutorial.collab.ipynb.\nConverted 50_tutorial.datablock.ipynb.\nConverted 60_medical.imaging.ipynb.\nConverted 61_tutorial.medical_imaging.ipynb.\nConverted 65_medical.text.ipynb.\nConverted 70_callback.wandb.ipynb.\nConverted 71_callback.tensorboard.ipynb.\nConverted 72_callback.neptune.ipynb.\nConverted 73_callback.captum.ipynb.\nConverted 74_callback.cutmix.ipynb.\nConverted 97_test_utils.ipynb.\nConverted 99_pytorch_doc.ipynb.\nConverted index.ipynb.\nConverted tutorial.ipynb.\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d00035cf4f5a61a585acf0b2f163831e7a3d6c66 | 97,108 | ipynb | Jupyter Notebook | notebooks/spark/other_notebooks/AerospikeSparkMLLinearRegression.ipynb | artanderson/interactive-notebooks | 73a4744eeabe53dfdfeb6a97d72d3969f9389700 | [
"MIT"
] | 11 | 2020-09-28T08:00:57.000Z | 2021-07-21T01:40:08.000Z | notebooks/spark/other_notebooks/AerospikeSparkMLLinearRegression.ipynb | artanderson/interactive-notebooks | 73a4744eeabe53dfdfeb6a97d72d3969f9389700 | [
"MIT"
] | 19 | 2020-10-02T16:35:32.000Z | 2022-02-12T22:46:04.000Z | notebooks/spark/other_notebooks/AerospikeSparkMLLinearRegression.ipynb | artanderson/interactive-notebooks | 73a4744eeabe53dfdfeb6a97d72d3969f9389700 | [
"MIT"
] | 17 | 2020-09-29T16:55:38.000Z | 2022-03-22T15:03:10.000Z | 104.305048 | 13,864 | 0.779112 | [
[
[
"# Aerospike Connect for Spark - SparkML Prediction Model Tutorial\n## Tested with Java 8, Spark 3.0.0, Python 3.7, and Aerospike Spark Connector 3.0.0",
"_____no_output_____"
],
[
"## Summary\nBuild a linear regression model to predict birth weight using Aerospike Database and Spark.\nHere are the features used:\n- gestation weeks\n- mother’s age\n- father’s age\n- mother’s weight gain during pregnancy\n- [Apgar score](https://en.wikipedia.org/wiki/Apgar_score)\n\nAerospike is used to store the Natality dataset that is published by CDC. The table is accessed in Apache Spark using the Aerospike Spark Connector, and Spark ML is used to build and evaluate the model. The model can later be converted to PMML and deployed on your inference server for predictions.",
"_____no_output_____"
],
[
"### Prerequisites\n\n1. Load Aerospike server if not alrady available - docker run -d --name aerospike -p 3000:3000 -p 3001:3001 -p 3002:3002 -p 3003:3003 aerospike\n2. Feature key needs to be located in AS_FEATURE_KEY_PATH\n3. [Download the connector](https://www.aerospike.com/enterprise/download/connectors/aerospike-spark/3.0.0/)",
"_____no_output_____"
]
],
[
[
"#IP Address or DNS name for one host in your Aerospike cluster. \n#A seed address for the Aerospike database cluster is required\nAS_HOST =\"127.0.0.1\"\n# Name of one of your namespaces. Type 'show namespaces' at the aql prompt if you are not sure\nAS_NAMESPACE = \"test\" \nAS_FEATURE_KEY_PATH = \"/etc/aerospike/features.conf\"\nAEROSPIKE_SPARK_JAR_VERSION=\"3.0.0\"\n\nAS_PORT = 3000 # Usually 3000, but change here if not\nAS_CONNECTION_STRING = AS_HOST + \":\"+ str(AS_PORT)",
"_____no_output_____"
],
[
"#Locate the Spark installation - this'll use the SPARK_HOME environment variable\n\nimport findspark\nfindspark.init()",
"_____no_output_____"
],
[
"# Below will help you download the Spark Connector Jar if you haven't done so already.\nimport urllib\nimport os\n\ndef aerospike_spark_jar_download_url(version=AEROSPIKE_SPARK_JAR_VERSION):\n DOWNLOAD_PREFIX=\"https://www.aerospike.com/enterprise/download/connectors/aerospike-spark/\"\n DOWNLOAD_SUFFIX=\"/artifact/jar\"\n AEROSPIKE_SPARK_JAR_DOWNLOAD_URL = DOWNLOAD_PREFIX+AEROSPIKE_SPARK_JAR_VERSION+DOWNLOAD_SUFFIX\n return AEROSPIKE_SPARK_JAR_DOWNLOAD_URL\n\ndef download_aerospike_spark_jar(version=AEROSPIKE_SPARK_JAR_VERSION):\n JAR_NAME=\"aerospike-spark-assembly-\"+AEROSPIKE_SPARK_JAR_VERSION+\".jar\"\n if(not(os.path.exists(JAR_NAME))) :\n urllib.request.urlretrieve(aerospike_spark_jar_download_url(),JAR_NAME)\n else :\n print(JAR_NAME+\" already downloaded\")\n return os.path.join(os.getcwd(),JAR_NAME)\n\nAEROSPIKE_JAR_PATH=download_aerospike_spark_jar()\nos.environ[\"PYSPARK_SUBMIT_ARGS\"] = '--jars ' + AEROSPIKE_JAR_PATH + ' pyspark-shell'",
"aerospike-spark-assembly-3.0.0.jar already downloaded\n"
],
[
"import pyspark\nfrom pyspark.context import SparkContext\nfrom pyspark.sql.context import SQLContext\nfrom pyspark.sql.session import SparkSession\nfrom pyspark.ml.linalg import Vectors\nfrom pyspark.ml.regression import LinearRegression\nfrom pyspark.sql.types import StringType, StructField, StructType, ArrayType, IntegerType, MapType, LongType, DoubleType",
"_____no_output_____"
],
[
"#Get a spark session object and set required Aerospike configuration properties\nsc = SparkContext.getOrCreate()\nprint(\"Spark Verison:\", sc.version)\n\nspark = SparkSession(sc)\nsqlContext = SQLContext(sc)\n\nspark.conf.set(\"aerospike.namespace\",AS_NAMESPACE)\nspark.conf.set(\"aerospike.seedhost\",AS_CONNECTION_STRING)\nspark.conf.set(\"aerospike.keyPath\",AS_FEATURE_KEY_PATH )",
"Spark Verison: 3.0.0\n"
]
],
[
[
"## Step 1: Load Data into a DataFrame",
"_____no_output_____"
]
],
[
[
"as_data=spark \\\n.read \\\n.format(\"aerospike\") \\\n.option(\"aerospike.set\", \"natality\").load()\n\nas_data.show(5)\n\nprint(\"Inferred Schema along with Metadata.\")\nas_data.printSchema()",
"+-----+--------------------+---------+------------+-------+-------------+---------------+-------------+----------+----------+----------+\n|__key| __digest| __expiry|__generation| __ttl| weight_pnd|weight_gain_pnd|gstation_week|apgar_5min|mother_age|father_age|\n+-----+--------------------+---------+------------+-------+-------------+---------------+-------------+----------+----------+----------+\n| null|[00 E0 68 A0 09 5...|354071840| 1|2367835| 6.9996768185| 99| 36| 99| 13| 15|\n| null|[01 B0 1F 4D D6 9...|354071839| 1|2367834| 5.291094288| 18| 40| 9| 14| 99|\n| null|[02 C0 93 23 F1 1...|354071837| 1|2367832| 6.8122838958| 24| 39| 9| 42| 36|\n| null|[02 B0 C4 C7 3B F...|354071838| 1|2367833|7.67649596284| 99| 39| 99| 14| 99|\n| null|[02 70 2A 45 E4 2...|354071843| 1|2367838| 7.8594796403| 40| 39| 8| 13| 99|\n+-----+--------------------+---------+------------+-------+-------------+---------------+-------------+----------+----------+----------+\nonly showing top 5 rows\n\nInferred Schema along with Metadata.\nroot\n |-- __key: string (nullable = true)\n |-- __digest: binary (nullable = false)\n |-- __expiry: integer (nullable = false)\n |-- __generation: integer (nullable = false)\n |-- __ttl: integer (nullable = false)\n |-- weight_pnd: double (nullable = true)\n |-- weight_gain_pnd: long (nullable = true)\n |-- gstation_week: long (nullable = true)\n |-- apgar_5min: long (nullable = true)\n |-- mother_age: long (nullable = true)\n |-- father_age: long (nullable = true)\n\n"
]
],
[
[
"### To speed up the load process at scale, use the [knobs](https://www.aerospike.com/docs/connect/processing/spark/performance.html) available in the Aerospike Spark Connector. \nFor example, **spark.conf.set(\"aerospike.partition.factor\", 15 )** will map 4096 Aerospike partitions to 32K Spark partitions. <font color=red> (Note: Please configure this carefully based on the available resources (CPU threads) in your system.)</font>",
"_____no_output_____"
],
[
"## Step 2 - Prep data",
"_____no_output_____"
]
],
[
[
"# This Spark3.0 setting, if true, will turn on Adaptive Query Execution (AQE), which will make use of the \n# runtime statistics to choose the most efficient query execution plan. It will speed up any joins that you\n# plan to use for data prep step.\nspark.conf.set(\"spark.sql.adaptive.enabled\", 'true')\n\n# Run a query in Spark SQL to ensure no NULL values exist.\nas_data.createOrReplaceTempView(\"natality\")\n\nsql_query = \"\"\"\nSELECT *\nfrom natality\nwhere weight_pnd is not null\nand mother_age is not null\nand father_age is not null\nand father_age < 80\nand gstation_week is not null\nand weight_gain_pnd < 90\nand apgar_5min != \"99\"\nand apgar_5min != \"88\"\n\"\"\"\nclean_data = spark.sql(sql_query)\n\n#Drop the Aerospike metadata from the dataset because its not required. \n#The metadata is added because we are inferring the schema as opposed to providing a strict schema\ncolumns_to_drop = ['__key','__digest','__expiry','__generation','__ttl' ]\nclean_data = clean_data.drop(*columns_to_drop)\n\n# dropping null values\nclean_data = clean_data.dropna()\n\n\nclean_data.cache()\nclean_data.show(5)\n\n#Descriptive Analysis of the data\nclean_data.describe().toPandas().transpose()",
"+------------------+---------------+-------------+----------+----------+----------+\n| weight_pnd|weight_gain_pnd|gstation_week|apgar_5min|mother_age|father_age|\n+------------------+---------------+-------------+----------+----------+----------+\n| 7.5398093604| 38| 39| 9| 42| 41|\n| 7.3634395508| 25| 37| 9| 14| 18|\n| 7.06361087448| 26| 39| 9| 42| 28|\n|6.1244416383599996| 20| 37| 9| 44| 41|\n| 7.06361087448| 49| 38| 9| 14| 18|\n+------------------+---------------+-------------+----------+----------+----------+\nonly showing top 5 rows\n\n"
]
],
[
[
"## Step 3 Visualize Data",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport math\n\n\npdf = clean_data.toPandas()\n\n#Histogram - Father Age\npdf[['father_age']].plot(kind='hist',bins=10,rwidth=0.8)\nplt.xlabel('Fathers Age (years)',fontsize=12)\nplt.legend(loc=None)\nplt.style.use('seaborn-whitegrid')\nplt.show()\n\n'''\npdf[['mother_age']].plot(kind='hist',bins=10,rwidth=0.8)\nplt.xlabel('Mothers Age (years)',fontsize=12)\nplt.legend(loc=None)\nplt.style.use('seaborn-whitegrid')\nplt.show()\n'''\n\npdf[['weight_pnd']].plot(kind='hist',bins=10,rwidth=0.8)\nplt.xlabel('Babys Weight (Pounds)',fontsize=12)\nplt.legend(loc=None)\nplt.style.use('seaborn-whitegrid')\nplt.show()\n\npdf[['gstation_week']].plot(kind='hist',bins=10,rwidth=0.8)\nplt.xlabel('Gestation (Weeks)',fontsize=12)\nplt.legend(loc=None)\nplt.style.use('seaborn-whitegrid')\nplt.show()\n\npdf[['weight_gain_pnd']].plot(kind='hist',bins=10,rwidth=0.8)\nplt.xlabel('mother’s weight gain during pregnancy',fontsize=12)\nplt.legend(loc=None)\nplt.style.use('seaborn-whitegrid')\nplt.show()\n\n#Histogram - Apgar Score\nprint(\"Apgar Score: Scores of 7 and above are generally normal; 4 to 6, fairly low; and 3 and below are generally \\\nregarded as critically low and cause for immediate resuscitative efforts.\")\npdf[['apgar_5min']].plot(kind='hist',bins=10,rwidth=0.8)\nplt.xlabel('Apgar score',fontsize=12)\nplt.legend(loc=None)\nplt.style.use('seaborn-whitegrid')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Step 4 - Create Model\n\n**Steps used for model creation:**\n1. Split cleaned data into Training and Test sets\n2. Vectorize features on which the model will be trained\n3. Create a linear regression model (Choose any ML algorithm that provides the best fit for the given dataset)\n4. Train model (Although not shown here, you could use K-fold cross-validation and Grid Search to choose the best hyper-parameters for the model)\n5. Evaluate model",
"_____no_output_____"
]
],
[
[
"# Define a function that collects the features of interest\n# (mother_age, father_age, and gestation_weeks) into a vector.\n# Package the vector in a tuple containing the label (`weight_pounds`) for that\n# row.## \n\ndef vector_from_inputs(r):\n return (r[\"weight_pnd\"], Vectors.dense(float(r[\"mother_age\"]),\n float(r[\"father_age\"]),\n float(r[\"gstation_week\"]),\n float(r[\"weight_gain_pnd\"]),\n float(r[\"apgar_5min\"])))\n\n\n",
"_____no_output_____"
],
[
"#Split that data 70% training and 30% Evaluation data\ntrain, test = clean_data.randomSplit([0.7, 0.3])\n\n#Check the shape of the data\ntrain.show()\nprint((train.count(), len(train.columns)))\ntest.show()\nprint((test.count(), len(test.columns)))",
"+------------------+---------------+-------------+----------+----------+----------+\n| weight_pnd|weight_gain_pnd|gstation_week|apgar_5min|mother_age|father_age|\n+------------------+---------------+-------------+----------+----------+----------+\n| 4.0565056208| 50| 33| 9| 44| 41|\n| 4.68702769012| 70| 36| 9| 44| 40|\n| 4.87442061282| 23| 33| 9| 43| 46|\n|6.1244416383599996| 20| 37| 9| 44| 41|\n|6.2501051276999995| 12| 38| 9| 44| 45|\n| 6.56316153974| 40| 38| 9| 47| 45|\n| 6.7681914434| 33| 39| 10| 47| 45|\n| 6.87621795178| 19| 38| 9| 44| 46|\n| 7.06361087448| 26| 39| 9| 42| 28|\n| 7.1099079495| 35| 39| 10| 43| 61|\n| 7.24879917456| 40| 37| 9| 44| 44|\n| 7.5398093604| 38| 39| 9| 42| 41|\n| 7.5618555866| 50| 38| 9| 42| 35|\n| 7.7492485093| 40| 38| 9| 44| 48|\n| 7.87491199864| 59| 41| 9| 43| 46|\n| 8.18796841068| 22| 40| 9| 42| 34|\n| 9.31232594688| 28| 41| 9| 45| 44|\n| 4.5856150496| 23| 36| 9| 42| 43|\n| 5.1257475915| 25| 36| 9| 54| 54|\n| 5.3131405142| 55| 36| 9| 47| 45|\n+------------------+---------------+-------------+----------+----------+----------+\nonly showing top 20 rows\n\n(5499, 6)\n+------------------+---------------+-------------+----------+----------+----------+\n| weight_pnd|weight_gain_pnd|gstation_week|apgar_5min|mother_age|father_age|\n+------------------+---------------+-------------+----------+----------+----------+\n| 3.62439958728| 50| 35| 9| 42| 37|\n| 5.3351867404| 6| 38| 9| 43| 48|\n| 6.8122838958| 24| 39| 9| 42| 36|\n| 6.9776305923| 27| 39| 9| 46| 42|\n| 7.06361087448| 49| 38| 9| 14| 18|\n| 7.3634395508| 25| 37| 9| 14| 18|\n| 7.4075320032| 18| 38| 9| 45| 45|\n| 7.68751907594| 25| 38| 10| 42| 49|\n| 3.09088091324| 42| 32| 9| 43| 46|\n| 5.62619692624| 24| 39| 9| 44| 50|\n|6.4992274837599995| 20| 39| 9| 42| 47|\n|6.5918216337999995| 63| 35| 9| 42| 38|\n| 6.686620406459999| 36| 38| 10| 14| 17|\n| 6.6910296517| 37| 40| 9| 42| 42|\n| 6.8122838958| 13| 35| 9| 14| 15|\n| 7.1870697412| 40| 36| 8| 14| 15|\n| 7.4075320032| 19| 40| 9| 43| 45|\n| 7.4736706818| 41| 37| 9| 43| 53|\n| 7.62578964258| 35| 38| 8| 43| 46|\n| 7.62578964258| 39| 39| 9| 42| 37|\n+------------------+---------------+-------------+----------+----------+----------+\nonly showing top 20 rows\n\n(2398, 6)\n"
],
[
"# Create an input DataFrame for Spark ML using the above function.\ntraining_data = train.rdd.map(vector_from_inputs).toDF([\"label\",\n \"features\"])\n \n# Construct a new LinearRegression object and fit the training data.\nlr = LinearRegression(maxIter=5, regParam=0.2, solver=\"normal\")\n\n#Voila! your first model using Spark ML is trained\nmodel = lr.fit(training_data)\n\n# Print the model summary.\nprint(\"Coefficients:\" + str(model.coefficients))\nprint(\"Intercept:\" + str(model.intercept))\nprint(\"R^2:\" + str(model.summary.r2))\nmodel.summary.residuals.show()",
"Coefficients:[0.00858931617782676,0.0008477851947958541,0.27948866120791893,0.009329081045860402,0.18817058385589935]\nIntercept:-5.893364345930709\nR^2:0.3970187134779115\n+--------------------+\n| residuals|\n+--------------------+\n| -1.845934264937739|\n| -2.2396120149639067|\n| -0.7717836944756593|\n| -0.6160804608336026|\n| -0.6986641251138215|\n| -0.672589930891391|\n| -0.8699157049741881|\n|-0.13870265354963962|\n|-0.26366319351660383|\n| -0.5260646593713352|\n| 0.3191520988648042|\n| 0.08956511232072462|\n| 0.28423773834709554|\n| 0.5367216316177004|\n|-0.34304851596998454|\n| 0.613435294490146|\n| 1.3680838827256254|\n| -1.887922569557201|\n| -1.4788456210255978|\n| -1.5035698497034602|\n+--------------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"### Evaluate Model",
"_____no_output_____"
]
],
[
[
"eval_data = test.rdd.map(vector_from_inputs).toDF([\"label\",\n \"features\"])\n\neval_data.show()\n\nevaluation_summary = model.evaluate(eval_data)\n\n\nprint(\"MAE:\", evaluation_summary.meanAbsoluteError)\nprint(\"RMSE:\", evaluation_summary.rootMeanSquaredError)\nprint(\"R-squared value:\", evaluation_summary.r2)",
"+------------------+--------------------+\n| label| features|\n+------------------+--------------------+\n| 3.62439958728|[42.0,37.0,35.0,5...|\n| 5.3351867404|[43.0,48.0,38.0,6...|\n| 6.8122838958|[42.0,36.0,39.0,2...|\n| 6.9776305923|[46.0,42.0,39.0,2...|\n| 7.06361087448|[14.0,18.0,38.0,4...|\n| 7.3634395508|[14.0,18.0,37.0,2...|\n| 7.4075320032|[45.0,45.0,38.0,1...|\n| 7.68751907594|[42.0,49.0,38.0,2...|\n| 3.09088091324|[43.0,46.0,32.0,4...|\n| 5.62619692624|[44.0,50.0,39.0,2...|\n|6.4992274837599995|[42.0,47.0,39.0,2...|\n|6.5918216337999995|[42.0,38.0,35.0,6...|\n| 6.686620406459999|[14.0,17.0,38.0,3...|\n| 6.6910296517|[42.0,42.0,40.0,3...|\n| 6.8122838958|[14.0,15.0,35.0,1...|\n| 7.1870697412|[14.0,15.0,36.0,4...|\n| 7.4075320032|[43.0,45.0,40.0,1...|\n| 7.4736706818|[43.0,53.0,37.0,4...|\n| 7.62578964258|[43.0,46.0,38.0,3...|\n| 7.62578964258|[42.0,37.0,39.0,3...|\n+------------------+--------------------+\nonly showing top 20 rows\n\nMAE: 0.9094828902906563\nRMSE: 1.1665322992147173\nR-squared value: 0.378390902740944\n"
]
],
[
[
"## Step 5 - Batch Prediction",
"_____no_output_____"
]
],
[
[
"#eval_data contains the records (ideally production) that you'd like to use for the prediction\n\npredictions = model.transform(eval_data)\npredictions.show()",
"+------------------+--------------------+-----------------+\n| label| features| prediction|\n+------------------+--------------------+-----------------+\n| 3.62439958728|[42.0,37.0,35.0,5...|6.440847435018738|\n| 5.3351867404|[43.0,48.0,38.0,6...| 6.88674880594522|\n| 6.8122838958|[42.0,36.0,39.0,2...|7.315398187463249|\n| 6.9776305923|[46.0,42.0,39.0,2...|7.382829406480911|\n| 7.06361087448|[14.0,18.0,38.0,4...|7.013375565916365|\n| 7.3634395508|[14.0,18.0,37.0,2...|6.509988959607797|\n| 7.4075320032|[45.0,45.0,38.0,1...|7.013333055266812|\n| 7.68751907594|[42.0,49.0,38.0,2...|7.244430398689434|\n| 3.09088091324|[43.0,46.0,32.0,4...|5.543968185959089|\n| 5.62619692624|[44.0,50.0,39.0,2...|7.344445812546044|\n|6.4992274837599995|[42.0,47.0,39.0,2...|7.287407500422561|\n|6.5918216337999995|[42.0,38.0,35.0,6...| 6.56297327380972|\n| 6.686620406459999|[14.0,17.0,38.0,3...|7.079420310981281|\n| 6.6910296517|[42.0,42.0,40.0,3...|7.721251613436126|\n| 6.8122838958|[14.0,15.0,35.0,1...|5.836519309057246|\n| 7.1870697412|[14.0,15.0,36.0,4...|6.179722574647495|\n| 7.4075320032|[43.0,45.0,40.0,1...|7.564460826372854|\n| 7.4736706818|[43.0,53.0,37.0,4...|6.938016907316393|\n| 7.62578964258|[43.0,46.0,38.0,3...| 6.96742600202968|\n| 7.62578964258|[42.0,37.0,39.0,3...|7.456182188345951|\n+------------------+--------------------+-----------------+\nonly showing top 20 rows\n\n"
]
],
[
[
"#### Compare the labels and the predictions, they should ideally match up for an accurate model. Label is the actual weight of the baby and prediction is the predicated weight",
"_____no_output_____"
],
[
"### Saving the Predictions to Aerospike for ML Application's consumption",
"_____no_output_____"
]
],
[
[
"# Aerospike is a key/value database, hence a key is needed to store the predictions into the database. Hence we need \n# to add the _id column to the predictions using SparkSQL\n\npredictions.createOrReplaceTempView(\"predict_view\")\n \nsql_query = \"\"\"\nSELECT *, monotonically_increasing_id() as _id\nfrom predict_view\n\"\"\"\npredict_df = spark.sql(sql_query)\npredict_df.show()\nprint(\"#records:\", predict_df.count())",
"+------------------+--------------------+-----------------+----------+\n| label| features| prediction| _id|\n+------------------+--------------------+-----------------+----------+\n| 3.62439958728|[42.0,37.0,35.0,5...|6.440847435018738| 0|\n| 5.3351867404|[43.0,48.0,38.0,6...| 6.88674880594522| 1|\n| 6.8122838958|[42.0,36.0,39.0,2...|7.315398187463249| 2|\n| 6.9776305923|[46.0,42.0,39.0,2...|7.382829406480911| 3|\n| 7.06361087448|[14.0,18.0,38.0,4...|7.013375565916365| 4|\n| 7.3634395508|[14.0,18.0,37.0,2...|6.509988959607797| 5|\n| 7.4075320032|[45.0,45.0,38.0,1...|7.013333055266812| 6|\n| 7.68751907594|[42.0,49.0,38.0,2...|7.244430398689434| 7|\n| 3.09088091324|[43.0,46.0,32.0,4...|5.543968185959089|8589934592|\n| 5.62619692624|[44.0,50.0,39.0,2...|7.344445812546044|8589934593|\n|6.4992274837599995|[42.0,47.0,39.0,2...|7.287407500422561|8589934594|\n|6.5918216337999995|[42.0,38.0,35.0,6...| 6.56297327380972|8589934595|\n| 6.686620406459999|[14.0,17.0,38.0,3...|7.079420310981281|8589934596|\n| 6.6910296517|[42.0,42.0,40.0,3...|7.721251613436126|8589934597|\n| 6.8122838958|[14.0,15.0,35.0,1...|5.836519309057246|8589934598|\n| 7.1870697412|[14.0,15.0,36.0,4...|6.179722574647495|8589934599|\n| 7.4075320032|[43.0,45.0,40.0,1...|7.564460826372854|8589934600|\n| 7.4736706818|[43.0,53.0,37.0,4...|6.938016907316393|8589934601|\n| 7.62578964258|[43.0,46.0,38.0,3...| 6.96742600202968|8589934602|\n| 7.62578964258|[42.0,37.0,39.0,3...|7.456182188345951|8589934603|\n+------------------+--------------------+-----------------+----------+\nonly showing top 20 rows\n\n#records: 2398\n"
],
[
"# Now we are good to write the Predictions to Aerospike\npredict_df \\\n.write \\\n.mode('overwrite') \\\n.format(\"aerospike\") \\\n.option(\"aerospike.writeset\", \"predictions\")\\\n.option(\"aerospike.updateByKey\", \"_id\") \\\n.save()",
"_____no_output_____"
]
],
[
[
"#### You can verify that data is written to Aerospike by using either [AQL](https://www.aerospike.com/docs/tools/aql/data_management.html) or the [Aerospike Data Browser](https://github.com/aerospike/aerospike-data-browser)",
"_____no_output_____"
],
[
"## Step 6 - Deploy\n### Here are a few options:\n1. Save the model to a PMML file by converting it using Jpmml/[pyspark2pmml](https://github.com/jpmml/pyspark2pmml) and load it into your production enviornment for inference.\n2. Use Aerospike as an [edge database for high velocity ingestion](https://medium.com/aerospike-developer-blog/add-horsepower-to-ai-ml-pipeline-15ca42a10982) for your inference pipline.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d0003cbcd9d17d2c4f06cd138b1bd9560704a09d | 30,840 | ipynb | Jupyter Notebook | notebook/fluent_ch18.ipynb | Lin0818/py-study-notebook | 6f70ab9a7fde0d6b46cd65475293e2eef6ef20e7 | [
"Apache-2.0"
] | 1 | 2018-12-12T09:00:27.000Z | 2018-12-12T09:00:27.000Z | notebook/fluent_ch18.ipynb | Lin0818/py-study-notebook | 6f70ab9a7fde0d6b46cd65475293e2eef6ef20e7 | [
"Apache-2.0"
] | null | null | null | notebook/fluent_ch18.ipynb | Lin0818/py-study-notebook | 6f70ab9a7fde0d6b46cd65475293e2eef6ef20e7 | [
"Apache-2.0"
] | null | null | null | 53.541667 | 1,424 | 0.6 | [
[
[
"## Concurrency with asyncio\n\n### Thread vs. coroutine\n",
"_____no_output_____"
]
],
[
[
"# spinner_thread.py\nimport threading \nimport itertools\nimport time\nimport sys\n\nclass Signal:\n go = True\n\ndef spin(msg, signal):\n write, flush = sys.stdout.write, sys.stdout.flush\n for char in itertools.cycle('|/-\\\\'):\n status = char + ' ' + msg\n write(status)\n flush()\n write('\\x08' * len(status))\n time.sleep(.1)\n if not signal.go:\n break\n write(' ' * len(status) + '\\x08' * len(status))\n\ndef slow_function():\n time.sleep(3)\n return 42\n\ndef supervisor():\n signal = Signal()\n spinner = threading.Thread(target=spin, args=('thinking!', signal))\n print('spinner object:', spinner)\n spinner.start()\n result = slow_function()\n signal.go = False\n spinner.join()\n return result\n\ndef main():\n result = supervisor()\n print('Answer:', result)\n \nif __name__ == '__main__':\n main()",
"spinner object: <Thread(Thread-6, initial)>\n| thinking/ thinking- thinking\\ thinking| thinking/ thinking- thinking\\ thinking| thinking/ thinking- thinking\\ thinking| thinking/ thinking- thinking\\ thinking| thinking/ thinking- thinking\\ thinking| thinking/ thinking- thinking\\ thinking| thinking/ thinking- thinking\\ thinking| thinking/ thinking Answer: 42\n"
],
[
"# spinner_asyncio.py\nimport asyncio\nimport itertools\nimport sys\n\[email protected]\ndef spin(msg):\n write, flush = sys.stdout.write, sys.stdout.flush\n for char in itertools.cycle('|/-\\\\'):\n status = char + ' ' + msg\n write(status)\n flush()\n write('\\x08' * len(status))\n try:\n yield from asyncio.sleep(.1)\n except asyncio.CancelledError:\n break\n write(' ' * len(status) + '\\x08' * len(status))\n \[email protected]\ndef slow_function():\n yield from asyncio.sleep(3)\n return 42\n\[email protected]\ndef supervisor():\n #Schedule the execution of a coroutine object: \n #wrap it in a future. Return a Task object.\n spinner = asyncio.ensure_future(spin('thinking!')) \n print('spinner object:', spinner)\n result = yield from slow_function()\n spinner.cancel()\n return result\n\ndef main():\n loop = asyncio.get_event_loop()\n result = loop.run_until_complete(supervisor())\n loop.close()\n print('Answer:', result)\n \nif __name__ == '__main__':\n main()",
"_____no_output_____"
],
[
"# flags_asyncio.py \nimport asyncio\n\nimport aiohttp\n\nfrom flags import BASE_URL, save_flag, show, main\n\[email protected]\ndef get_flag(cc):\n url = '{}/{cc}/{cc}.gif'.format(BASE_URL, cc=cc.lower())\n resp = yield from aiohttp.request('GET', url)\n image = yield from resp.read()\n return image\n\[email protected]\ndef download_one(cc):\n image = yield from get_flag(cc)\n show(cc)\n save_flag(image, cc.lower() + '.gif')\n return cc\n\ndef download_many(cc_list):\n loop = asyncio.get_event_loop()\n to_do = [download_one(cc) for cc in sorted(cc_list)]\n wait_coro = asyncio.wait(to_do)\n res, _ = loop.run_until_complete(wait_coro)\n loop.close()\n \n return len(res)\n\nif __name__ == '__main__':\n main(download_many)",
"_____no_output_____"
],
[
"# flags2_asyncio.py\nimport asyncio\nimport collections\n\nimport aiohttp\nfrom aiohttp import web\nimport tqdm \n\nfrom flags2_common import HTTPStatus, save_flag, Result, main\n\nDEFAULT_CONCUR_REQ = 5\nMAX_CONCUR_REQ = 1000\n\nclass FetchError(Exception):\n def __init__(self, country_code):\n self.country_code = country_code\n\[email protected]\ndef get_flag(base_url, cc):\n url = '{}/{cc}/{cc}.gif'.format(BASE_URL, cc=cc.lower())\n resp = yield from aiohttp.ClientSession().get(url)\n if resp.status == 200:\n image = yield from resp.read()\n return image\n elif resp.status == 404:\n raise web.HTTPNotFound()\n else:\n raise aiohttp.HttpProcessingError(\n code=resp.status, message=resp.reason, headers=resp.headers)\n\[email protected] \ndef download_one(cc, base_url, semaphore, verbose):\n try:\n with (yield from semaphore):\n image = yield from get_flag(base_url, cc)\n except web.HTTPNotFound:\n status = HTTPStatus.not_found \n msg = 'not found'\n except Exception as exc:\n raise FetchError(cc) from exc\n else:\n save_flag(image, cc.lower() + '.gif') \n status = HTTPStatus.ok\n msg = 'OK'\n if verbose and msg: \n print(cc, msg)\n \n return Result(status, cc)\n\[email protected]\ndef downloader_coro(cc_list, base_url, verbose, concur_req): \n counter = collections.Counter()\n semaphore = asyncio.Semaphore(concur_req)\n to_do = [download_one(cc, base_url, semaphore, verbose)\n for cc in sorted(cc_list)]\n to_do_iter = asyncio.as_completed(to_do) \n if not verbose:\n to_do_iter = tqdm.tqdm(to_do_iter, total=len(cc_list)) \n for future in to_do_iter:\n try:\n res = yield from future\n except FetchError as exc: \n country_code = exc.country_code \n try:\n error_msg = exc.__cause__.args[0] \n except IndexError:\n error_msg = exc.__cause__.__class__.__name__ \n if verbose and error_msg:\n msg = '*** Error for {}: {}'\n print(msg.format(country_code, error_msg)) \n status = HTTPStatus.error\n else:\n status = res.status\n counter[status] += 1 \n return counter\n\ndef download_many(cc_list, base_url, verbose, concur_req):\n loop = asyncio.get_event_loop()\n coro = download_coro(cc_list, base_url, verbose, concur_req)\n counts = loop.run_until_complete(wait_coro)\n loop.close()\n\n return counts\n\nif __name__ == '__main__':\n main(download_many, DEFAULT_CONCUR_REQ, MAX_CONCUR_REQ)",
"_____no_output_____"
],
[
"# run_in_executor\[email protected]\ndef download_one(cc, base_url, semaphore, verbose):\n try:\n with (yield from semaphore):\n image = yield from get_flag(base_url, cc)\n except web.HTTPNotFound:\n status = HTTPStatus.not_found\n msg = 'not found'\n except Exception as exc:\n raise FetchError(cc) from exc\n else:\n # save_flag 也是阻塞操作,所以使用run_in_executor调用save_flag进行\n # 异步操作\n loop = asyncio.get_event_loop()\n loop.run_in_executor(None, save_flag, image, cc.lower() + '.gif')\n status = HTTPStatus.ok\n msg = 'OK'\n \n if verbose and msg:\n print(cc, msg)\n \n return Result(status, cc)",
"_____no_output_____"
],
[
"## Doing multiple requests for each download\n# flags3_asyncio.py\[email protected]\ndef http_get(url):\n res = yield from aiohttp.request('GET', url)\n if res.status == 200:\n ctype = res.headers.get('Content-type', '').lower()\n if 'json' in ctype or url.endswith('json'):\n data = yield from res.json()\n else:\n data = yield from res.read()\n \n elif res.status == 404:\n raise web.HTTPNotFound()\n else:\n raise aiohttp.errors.HttpProcessingError(\n code=res.status, message=res.reason,\n headers=res.headers)\n \[email protected]\ndef get_country(base_url, cc):\n url = '{}/{cc}/metadata.json'.format(base_url, cc=cc.lower())\n metadata = yield from http_get(url)\n return metadata['country']\n\[email protected]\ndef get_flag(base_url, cc):\n url = '{}/{cc}/{cc}.gif'.format(base_url, cc=cc.lower())\n return (yield from http_get(url))\n\[email protected]\ndef download_one(cc, base_url, semaphore, verbose):\n try:\n with (yield from semaphore):\n image = yield from get_flag(base_url, cc)\n with (yield from semaphore):\n country = yield from get_country(base_url, cc)\n except web.HTTPNotFound:\n status = HTTPStatus.not_found\n msg = 'not found'\n except Exception as exc:\n raise FetchError(cc) from exc\n else:\n country = country.replace(' ', '_')\n filename = '{}-{}.gif'.format(country, cc)\n loop = asyncio.get_event_loop()\n loop.run_in_executor(None, save_flag, image, filename)\n status = HTTPStatus.ok\n msg = 'OK'\n \n if verbose and msg:\n print(cc, msg)\n \n return Result(status, cc)",
"_____no_output_____"
]
],
[
[
"### Writing asyncio servers",
"_____no_output_____"
]
],
[
[
"# tcp_charfinder.py\nimport sys\nimport asyncio\n\nfrom charfinder import UnicodeNameIndex\n\nCRLF = b'\\r\\n'\nPROMPT = b'?>'\n\nindex = UnicodeNameIndex()\n\[email protected]\ndef handle_queries(reader, writer):\n while True:\n writer.write(PROMPT)\n yield from writer.drain()\n data = yield from reader.readline()\n try:\n query = data.decode().strip()\n except UnicodeDecodeError:\n query = '\\x00'\n client = writer.get_extra_info('peername')\n print('Received from {}: {!r}'.format(client, query))\n if query:\n if ord(query[:1]) < 32:\n break\n lines = list(index.find_description_strs(query))\n if lines:\n writer.writelines(line.encode() + CRLF for line in lines)\n writer.write(index.status(query, len(lines)).encode() + CRLF)\n \n yield from writer.drain()\n print('Sent {} results'.format(len(lines)))\n print('Close the client socket')\n writer.close()\n\ndef main(address='127.0.0.1', port=2323):\n port = int(port)\n loop = asyncio.get_event_loop()\n server_coro = asyncio.start_server(handle_queries, address, port, loop=loop)\n server = loop.run_until_complete(server_coro)\n \n host = server.sockets[0].getsockname()\n print('Serving on {}. Hit CTRL-C to stop.'.format(host))\n try:\n loop.run_forever()\n except KeyboardInterrupt:\n pass\n \n print('Server shutting down.')\n server.close()\n loop.run_until_complete(server.wait_closed())\n loop.close()\n \nif __name__ == '__main__':\n main()",
"_____no_output_____"
],
[
"# http_charfinder.py\[email protected]\ndef init(loop, address, port):\n app = web.Application(loop=loop)\n app.router.add_route('GET', '/', home)\n handler = app.make_handler()\n server = yield from loop.create_server(handler, address, port)\n return server.sockets[0].getsockname()\n\ndef home(request):\n query = request.GET.get('query', '').strip()\n print('Query: {!r}'.format(query))\n if query:\n descriptions = list(index.find_descriptions(query))\n res = '\\n'.join(ROW_TPL.format(**vars(descr)) \n for descr in descriptions)\n msg = index.status(query, len(descriptions))\n else:\n descriptions = []\n res = ''\n msg = 'Enter words describing characters.'\n \n html = template.format(query=query, result=res, message=msg)\n print('Sending {} results'.format(len(descriptions)))\n return web.Response(content_type=CONTENT_TYPE, text=html)\n \ndef main(address='127.0.0.1', port=8888):\n port = int(port)\n loop = asyncio.get_event_loop()\n host = loop.run_until_complete(init(loop, address, port))\n print('Serving on {}. Hit CTRL-C to stop.'.format(host))\n try:\n loop.run_forever()\n except KeyboardInterrupt: # CTRL+C pressed\n pass\n print('Server shutting down.')\n loop.close()\n \nif __name__ == '__main__':\n main(*sys.argv[1:])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d000491b7c790e6ee107777a67eb83691ed8c106 | 4,243 | ipynb | Jupyter Notebook | Sessions/Problem-1.ipynb | Yunika-Bajracharya/pybasics | e04a014b70262ef9905fef5720f58a6f0acc0fda | [
"CC-BY-4.0"
] | 1 | 2020-07-14T13:34:41.000Z | 2020-07-14T13:34:41.000Z | Sessions/Problem-1.ipynb | JahBirShakya/pybasics | e04a014b70262ef9905fef5720f58a6f0acc0fda | [
"CC-BY-4.0"
] | null | null | null | Sessions/Problem-1.ipynb | JahBirShakya/pybasics | e04a014b70262ef9905fef5720f58a6f0acc0fda | [
"CC-BY-4.0"
] | null | null | null | 40.409524 | 468 | 0.587556 | [
[
[
"## Problem 1\n---\n\n#### The solution should try to use all the python constructs\n\n- Conditionals and Loops\n- Functions\n- Classes\n\n#### and datastructures as possible\n\n- List\n- Tuple\n- Dictionary\n- Set",
"_____no_output_____"
],
[
"### Problem\n---\n\nMoist has a hobby -- collecting figure skating trading cards. His card collection has been growing, and it is now too large to keep in one disorganized pile. Moist needs to sort the cards in alphabetical order, so that he can find the cards that he wants on short notice whenever it is necessary.\n\nThe problem is -- Moist can't actually pick up the cards because they keep sliding out his hands, and the sweat causes permanent damage. Some of the cards are rather expensive, mind you. To facilitate the sorting, Moist has convinced Dr. Horrible to build him a sorting robot. However, in his rather horrible style, Dr. Horrible has decided to make the sorting robot charge Moist a fee of $1 whenever it has to move a trading card during the sorting process.\n\nMoist has figured out that the robot's sorting mechanism is very primitive. It scans the deck of cards from top to bottom. Whenever it finds a card that is lexicographically smaller than the previous card, it moves that card to its correct place in the stack above. This operation costs $1, and the robot resumes scanning down towards the bottom of the deck, moving cards one by one until the entire deck is sorted in lexicographical order from top to bottom.\n\nAs wet luck would have it, Moist is almost broke, but keeping his trading cards in order is the only remaining joy in his miserable life. He needs to know how much it would cost him to use the robot to sort his deck of cards.\nInput\n\nThe first line of the input gives the number of test cases, **T**. **T** test cases follow. Each one starts with a line containing a single integer, **N**. The next **N** lines each contain the name of a figure skater, in order from the top of the deck to the bottom.\nOutput\n\nFor each test case, output one line containing \"Case #x: y\", where x is the case number (starting from 1) and y is the number of dollars it would cost Moist to use the robot to sort his deck of trading cards.\nLimits\n\n1 ≤ **T** ≤ 100.\nEach name will consist of only letters and the space character.\nEach name will contain at most 100 characters.\nNo name with start or end with a space.\nNo name will appear more than once in the same test case.\nLexicographically, the space character comes first, then come the upper case letters, then the lower case letters.\n\nSmall dataset\n\n1 ≤ N ≤ 10.\n\nLarge dataset\n\n1 ≤ N ≤ 100.\n\nSample\n\n\n| Input | Output |\n|---------------------|-------------|\n| 2 | Case \\#1: 1 | \n| 2 | Case \\#2: 0 |\n| Oksana Baiul | |\n| Michelle Kwan | |\n| 3 | |\n| Elvis Stojko | |\n| Evgeni Plushenko | |\n| Kristi Yamaguchi | |\n\n\n\n*Note: Solution is not important but procedure taken to solve the problem is important*\n\t\n\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown"
]
] |
d0004ddbda5277669a00c9cb8161daa5a9dbecdb | 3,548 | ipynb | Jupyter Notebook | filePreprocessing.ipynb | zinccat/WeiboTextClassification | ec3729450f1aa0cfa2657cac955334cfae565047 | [
"MIT"
] | 2 | 2020-03-28T11:09:51.000Z | 2020-04-06T13:01:14.000Z | filePreprocessing.ipynb | zinccat/WeiboTextClassification | ec3729450f1aa0cfa2657cac955334cfae565047 | [
"MIT"
] | null | null | null | filePreprocessing.ipynb | zinccat/WeiboTextClassification | ec3729450f1aa0cfa2657cac955334cfae565047 | [
"MIT"
] | null | null | null | 27.937008 | 86 | 0.463641 | [
[
[
"### 原始数据处理程序",
"_____no_output_____"
],
[
"本程序用于将原始txt格式数据以utf-8编码写入到csv文件中, 以便后续操作\n\n请在使用前确认原始数据所在文件夹内无无关文件,并修改各分类文件夹名至1-9\n\n一个可行的对应关系如下所示:\n\n财经 1 economy\n房产 2 realestate\n健康 3 health\n教育 4 education\n军事 5 military\n科技 6 technology\n体育 7 sports\n娱乐 8 entertainment\n证券 9 stock",
"_____no_output_____"
],
[
"先导入一些库",
"_____no_output_____"
]
],
[
[
"import os #用于文件操作\nimport pandas as pd #用于读写数据",
"_____no_output_____"
]
],
[
[
"数据处理所用函数,读取文件夹名作为数据的类别,将数据以文本(text),类别(category)的形式输出至csv文件中\n\n传入参数: corpus_path: 原始语料库根目录 out_path: 处理后文件输出目录",
"_____no_output_____"
]
],
[
[
"def processing(corpus_path, out_path):\n if not os.path.exists(out_path): #检测输出目录是否存在,若不存在则创建目录\n os.makedirs(out_path)\n clist = os.listdir(corpus_path) #列出原始数据根目录下的文件夹\n for classid in clist: #对每个文件夹分别处理\n dict = {'text': [], 'category': []}\n class_path = corpus_path+classid+\"/\"\n filelist = os.listdir(class_path)\n for fileN in filelist: #处理单个文件\n file_path = class_path + fileN\n with open(file_path, encoding='utf-8', errors='ignore') as f:\n content = f.read()\n dict['text'].append(content) #将文本内容加入字典\n dict['category'].append(classid) #将类别加入字典\n pf = pd.DataFrame(dict, columns=[\"text\", \"category\"])\n if classid == '1': #第一类数据输出时创建新文件并添加header\n pf.to_csv(out_path+'dataUTF8.csv', mode='w',\n header=True, encoding='utf-8', index=False)\n else: #将剩余类别的数据写入到已生成的文件中\n pf.to_csv(out_path+'dataUTF8.csv', mode='a',\n header=False, encoding='utf-8', index=False)",
"_____no_output_____"
]
],
[
[
"处理文件",
"_____no_output_____"
]
],
[
[
"processing(\"./data/\", \"./dataset/\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d00053774622cc4b262f99d26678120db756bf21 | 38,336 | ipynb | Jupyter Notebook | IBM_AI/4_Pytorch/5.1logistic_regression_prediction_v2.ipynb | merula89/cousera_notebooks | caa529a7abd3763d26f3f2add7c3ab508fbb9bd2 | [
"MIT"
] | null | null | null | IBM_AI/4_Pytorch/5.1logistic_regression_prediction_v2.ipynb | merula89/cousera_notebooks | caa529a7abd3763d26f3f2add7c3ab508fbb9bd2 | [
"MIT"
] | null | null | null | IBM_AI/4_Pytorch/5.1logistic_regression_prediction_v2.ipynb | merula89/cousera_notebooks | caa529a7abd3763d26f3f2add7c3ab508fbb9bd2 | [
"MIT"
] | null | null | null | 40.226653 | 8,660 | 0.717054 | [
[
[
"<a href=\"http://cocl.us/pytorch_link_top\">\n <img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DL0110EN/notebook_images%20/Pytochtop.png\" width=\"750\" alt=\"IBM Product \" />\n</a> ",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DL0110EN/notebook_images%20/cc-logo-square.png\" width=\"200\" alt=\"cognitiveclass.ai logo\" />",
"_____no_output_____"
],
[
"<h1>Logistic Regression</h1>",
"_____no_output_____"
],
[
"<h2>Table of Contents</h2>\n<p>In this lab, we will cover logistic regression using PyTorch.</p>\n\n<ul>\n <li><a href=\"#Log\">Logistic Function</a></li>\n <li><a href=\"#Seq\">Build a Logistic Regression Using nn.Sequential</a></li>\n <li><a href=\"#Model\">Build Custom Modules</a></li>\n</ul>\n<p>Estimated Time Needed: <strong>15 min</strong></p>\n\n<hr>",
"_____no_output_____"
],
[
"<h2>Preparation</h2>",
"_____no_output_____"
],
[
"We'll need the following libraries: ",
"_____no_output_____"
]
],
[
[
"# Import the libraries we need for this lab\n\nimport torch.nn as nn\nimport torch\nimport matplotlib.pyplot as plt ",
"_____no_output_____"
]
],
[
[
"Set the random seed:",
"_____no_output_____"
]
],
[
[
"# Set the random seed\n\ntorch.manual_seed(2)",
"_____no_output_____"
]
],
[
[
"<!--Empty Space for separating topics-->",
"_____no_output_____"
],
[
"<h2 id=\"Log\">Logistic Function</h2>",
"_____no_output_____"
],
[
"Create a tensor ranging from -100 to 100:",
"_____no_output_____"
]
],
[
[
"z = torch.arange(-100, 100, 0.1).view(-1, 1)\nprint(\"The tensor: \", z)",
"The tensor: tensor([[-100.0000],\n [ -99.9000],\n [ -99.8000],\n ...,\n [ 99.7000],\n [ 99.8000],\n [ 99.9000]])\n"
]
],
[
[
"Create a sigmoid object: ",
"_____no_output_____"
]
],
[
[
"# Create sigmoid object\n\nsig = nn.Sigmoid()",
"_____no_output_____"
]
],
[
[
"Apply the element-wise function Sigmoid with the object:",
"_____no_output_____"
]
],
[
[
"# Use sigmoid object to calculate the \n\nyhat = sig(z)",
"_____no_output_____"
]
],
[
[
"Plot the results: ",
"_____no_output_____"
]
],
[
[
"plt.plot(z.numpy(), yhat.numpy())\nplt.xlabel('z')\nplt.ylabel('yhat')",
"_____no_output_____"
]
],
[
[
"Apply the element-wise Sigmoid from the function module and plot the results:",
"_____no_output_____"
]
],
[
[
"yhat = torch.sigmoid(z)\nplt.plot(z.numpy(), yhat.numpy())",
"_____no_output_____"
]
],
[
[
"<!--Empty Space for separating topics-->",
"_____no_output_____"
],
[
"<h2 id=\"Seq\">Build a Logistic Regression with <code>nn.Sequential</code></h2>",
"_____no_output_____"
],
[
"Create a 1x1 tensor where x represents one data sample with one dimension, and 2x1 tensor X represents two data samples of one dimension:",
"_____no_output_____"
]
],
[
[
"# Create x and X tensor\n\nx = torch.tensor([[1.0]])\nX = torch.tensor([[1.0], [100]])\nprint('x = ', x)\nprint('X = ', X)",
"x = tensor([[1.]])\nX = tensor([[ 1.],\n [100.]])\n"
]
],
[
[
"Create a logistic regression object with the <code>nn.Sequential</code> model with a one-dimensional input:",
"_____no_output_____"
]
],
[
[
"# Use sequential function to create model\n\nmodel = nn.Sequential(nn.Linear(1, 1), nn.Sigmoid())",
"_____no_output_____"
]
],
[
[
"The object is represented in the following diagram: ",
"_____no_output_____"
],
[
"<img src = \"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DL0110EN/notebook_images%20/chapter3/3.1.1_logistic_regression_block_diagram.png\" width = 800, align = \"center\" alt=\"logistic regression block diagram\" />",
"_____no_output_____"
],
[
"In this case, the parameters are randomly initialized. You can view them the following ways:",
"_____no_output_____"
]
],
[
[
"# Print the parameters\n\nprint(\"list(model.parameters()):\\n \", list(model.parameters()))\nprint(\"\\nmodel.state_dict():\\n \", model.state_dict())",
"list(model.parameters()):\n [Parameter containing:\ntensor([[0.2294]], requires_grad=True), Parameter containing:\ntensor([-0.2380], requires_grad=True)]\n\nmodel.state_dict():\n OrderedDict([('0.weight', tensor([[0.2294]])), ('0.bias', tensor([-0.2380]))])\n"
]
],
[
[
"Make a prediction with one sample:",
"_____no_output_____"
]
],
[
[
"# The prediction for x\n\nyhat = model(x)\nprint(\"The prediction: \", yhat)",
"The prediction: tensor([[0.4979]], grad_fn=<SigmoidBackward>)\n"
]
],
[
[
"Calling the object with tensor <code>X</code> performed the following operation <b>(code values may not be the same as the diagrams value depending on the version of PyTorch) </b>:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DL0110EN/notebook_images%20/chapter3/3.1.1_logistic_functio_example%20.png\" width=\"400\" alt=\"Logistic Example\" />",
"_____no_output_____"
],
[
"Make a prediction with multiple samples:",
"_____no_output_____"
]
],
[
[
"# The prediction for X\n\nyhat = model(X)\nyhat",
"_____no_output_____"
]
],
[
[
"Calling the object performed the following operation: ",
"_____no_output_____"
],
[
"Create a 1x2 tensor where x represents one data sample with one dimension, and 2x3 tensor X represents one data sample of two dimensions:",
"_____no_output_____"
]
],
[
[
"# Create and print samples\n\nx = torch.tensor([[1.0, 1.0]])\nX = torch.tensor([[1.0, 1.0], [1.0, 2.0], [1.0, 3.0]])\nprint('x = ', x)\nprint('X = ', X)",
"x = tensor([[1., 1.]])\nX = tensor([[1., 1.],\n [1., 2.],\n [1., 3.]])\n"
]
],
[
[
"Create a logistic regression object with the <code>nn.Sequential</code> model with a two-dimensional input: ",
"_____no_output_____"
]
],
[
[
"# Create new model using nn.sequential()\n\nmodel = nn.Sequential(nn.Linear(2, 1), nn.Sigmoid())",
"_____no_output_____"
]
],
[
[
"The object will apply the Sigmoid function to the output of the linear function as shown in the following diagram:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DL0110EN/notebook_images%20/chapter3/3.1.1logistic_output.png\" width=\"800\" alt=\"The structure of nn.sequential\"/>",
"_____no_output_____"
],
[
"In this case, the parameters are randomly initialized. You can view them the following ways:",
"_____no_output_____"
]
],
[
[
"# Print the parameters\n\nprint(\"list(model.parameters()):\\n \", list(model.parameters()))\nprint(\"\\nmodel.state_dict():\\n \", model.state_dict())",
"list(model.parameters()):\n [Parameter containing:\ntensor([[ 0.1939, -0.0361]], requires_grad=True), Parameter containing:\ntensor([0.3021], requires_grad=True)]\n\nmodel.state_dict():\n OrderedDict([('0.weight', tensor([[ 0.1939, -0.0361]])), ('0.bias', tensor([0.3021]))])\n"
]
],
[
[
"Make a prediction with one sample:",
"_____no_output_____"
]
],
[
[
"# Make the prediction of x\n\nyhat = model(x)\nprint(\"The prediction: \", yhat)",
"The prediction: tensor([[0.6130]], grad_fn=<SigmoidBackward>)\n"
]
],
[
[
"The operation is represented in the following diagram:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DL0110EN/notebook_images%20/chapter3/3.3.1.logisticwithouptut.png\" width=\"500\" alt=\"Sequential Example\" />",
"_____no_output_____"
],
[
"Make a prediction with multiple samples:",
"_____no_output_____"
]
],
[
[
"# The prediction of X\n\nyhat = model(X)\nprint(\"The prediction: \", yhat)",
"_____no_output_____"
]
],
[
[
"The operation is represented in the following diagram: ",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DL0110EN/notebook_images%20/chapter3/3.1.1_logistic_with_outputs2.png\" width=\"800\" alt=\"Sequential Example\" />",
"_____no_output_____"
],
[
"<!--Empty Space for separating topics-->",
"_____no_output_____"
],
[
"<h2 id=\"Model\">Build Custom Modules</h2>",
"_____no_output_____"
],
[
"In this section, you will build a custom Module or class. The model or object function is identical to using <code>nn.Sequential</code>.",
"_____no_output_____"
],
[
"Create a logistic regression custom module:",
"_____no_output_____"
]
],
[
[
"# Create logistic_regression custom class\n\nclass logistic_regression(nn.Module):\n \n # Constructor\n def __init__(self, n_inputs):\n super(logistic_regression, self).__init__()\n self.linear = nn.Linear(n_inputs, 1)\n \n # Prediction\n def forward(self, x):\n yhat = torch.sigmoid(self.linear(x))\n return yhat",
"_____no_output_____"
]
],
[
[
"Create a 1x1 tensor where x represents one data sample with one dimension, and 3x1 tensor where $X$ represents one data sample of one dimension:",
"_____no_output_____"
]
],
[
[
"# Create x and X tensor\n\nx = torch.tensor([[1.0]])\nX = torch.tensor([[-100], [0], [100.0]])\nprint('x = ', x)\nprint('X = ', X)",
"_____no_output_____"
]
],
[
[
"Create a model to predict one dimension: ",
"_____no_output_____"
]
],
[
[
"# Create logistic regression model\n\nmodel = logistic_regression(1)",
"_____no_output_____"
]
],
[
[
"In this case, the parameters are randomly initialized. You can view them the following ways:",
"_____no_output_____"
]
],
[
[
"# Print parameters \n\nprint(\"list(model.parameters()):\\n \", list(model.parameters()))\nprint(\"\\nmodel.state_dict():\\n \", model.state_dict())",
"_____no_output_____"
]
],
[
[
"Make a prediction with one sample:",
"_____no_output_____"
]
],
[
[
"# Make the prediction of x\n\nyhat = model(x)\nprint(\"The prediction result: \\n\", yhat)",
"_____no_output_____"
]
],
[
[
"Make a prediction with multiple samples:",
"_____no_output_____"
]
],
[
[
"# Make the prediction of X\n\nyhat = model(X)\nprint(\"The prediction result: \\n\", yhat)",
"_____no_output_____"
]
],
[
[
"Create a logistic regression object with a function with two inputs: ",
"_____no_output_____"
]
],
[
[
"# Create logistic regression model\n\nmodel = logistic_regression(2)",
"_____no_output_____"
]
],
[
[
"Create a 1x2 tensor where x represents one data sample with one dimension, and 3x2 tensor X represents one data sample of one dimension:",
"_____no_output_____"
]
],
[
[
"# Create x and X tensor\n\nx = torch.tensor([[1.0, 2.0]])\nX = torch.tensor([[100, -100], [0.0, 0.0], [-100, 100]])\nprint('x = ', x)\nprint('X = ', X)",
"_____no_output_____"
]
],
[
[
"Make a prediction with one sample:",
"_____no_output_____"
]
],
[
[
"# Make the prediction of x\n\nyhat = model(x)\nprint(\"The prediction result: \\n\", yhat)",
"_____no_output_____"
]
],
[
[
"Make a prediction with multiple samples: ",
"_____no_output_____"
]
],
[
[
"# Make the prediction of X\n\nyhat = model(X)\nprint(\"The prediction result: \\n\", yhat)",
"_____no_output_____"
]
],
[
[
"<!--Empty Space for separating topics-->",
"_____no_output_____"
],
[
"<h3>Practice</h3>",
"_____no_output_____"
],
[
"Make your own model <code>my_model</code> as applying linear regression first and then logistic regression using <code>nn.Sequential()</code>. Print out your prediction.",
"_____no_output_____"
]
],
[
[
"# Practice: Make your model and make the prediction\n\nX = torch.tensor([-10.0])",
"_____no_output_____"
]
],
[
[
"Double-click <b>here</b> for the solution.\n\n<!-- \nmy_model = nn.Sequential(nn.Linear(1, 1),nn.Sigmoid())\nyhat = my_model(X)\nprint(\"The prediction: \", yhat)\n-->",
"_____no_output_____"
],
[
"<!--Empty Space for separating topics-->",
"_____no_output_____"
],
[
"<a href=\"http://cocl.us/pytorch_link_bottom\">\n <img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/DL0110EN/notebook_images%20/notebook_bottom%20.png\" width=\"750\" alt=\"PyTorch Bottom\" />\n</a>",
"_____no_output_____"
],
[
"<h2>About the Authors:</h2> \n\n<a href=\"https://www.linkedin.com/in/joseph-s-50398b136/\">Joseph Santarcangelo</a> has a PhD in Electrical Engineering, his research focused on using machine learning, signal processing, and computer vision to determine how videos impact human cognition. Joseph has been working for IBM since he completed his PhD. ",
"_____no_output_____"
],
[
"Other contributors: <a href=\"https://www.linkedin.com/in/michelleccarey/\">Michelle Carey</a>, <a href=\"www.linkedin.com/in/jiahui-mavis-zhou-a4537814a\">Mavis Zhou</a>",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"Copyright © 2018 <a href=\"cognitiveclass.ai?utm_source=bducopyrightlink&utm_medium=dswb&utm_campaign=bdu\">cognitiveclass.ai</a>. This notebook and its source code are released under the terms of the <a href=\"https://bigdatauniversity.com/mit-license/\">MIT License</a>.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0005af9eace679454187ce22b2c411130a19e72 | 1,217 | ipynb | Jupyter Notebook | Access Environment variable.ipynb | shkhaider2015/PIAIC-QUARTER-2 | 2b6ef1c8d75f9f52b9da8e735751f5f80c76b227 | [
"Unlicense"
] | null | null | null | Access Environment variable.ipynb | shkhaider2015/PIAIC-QUARTER-2 | 2b6ef1c8d75f9f52b9da8e735751f5f80c76b227 | [
"Unlicense"
] | null | null | null | Access Environment variable.ipynb | shkhaider2015/PIAIC-QUARTER-2 | 2b6ef1c8d75f9f52b9da8e735751f5f80c76b227 | [
"Unlicense"
] | null | null | null | 17.140845 | 59 | 0.506163 | [
[
[
"import os",
"_____no_output_____"
],
[
"db_user = os.environ.get('DB_USER')\ndb_user_password = os.environ.get('DB_USER_PASSWORD')",
"_____no_output_____"
],
[
"print(db_user)\nprint(db_user_password)",
"shkhaider2015\nProgressive0314\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code"
]
] |
d00070e01aa3101ac81e3c3f48915570e8611db3 | 5,451 | ipynb | Jupyter Notebook | stemming.ipynb | Ganeshatmuri/NaturalLanguageProcessing | 491d5bc50559c7a09e0b541a96c4314c20b80927 | [
"Unlicense"
] | null | null | null | stemming.ipynb | Ganeshatmuri/NaturalLanguageProcessing | 491d5bc50559c7a09e0b541a96c4314c20b80927 | [
"Unlicense"
] | null | null | null | stemming.ipynb | Ganeshatmuri/NaturalLanguageProcessing | 491d5bc50559c7a09e0b541a96c4314c20b80927 | [
"Unlicense"
] | null | null | null | 45.806723 | 1,257 | 0.613832 | [
[
[
"import nltk\nfrom nltk.stem import PorterStemmer\nfrom nltk.corpus import stopwords\nimport re",
"_____no_output_____"
],
[
"paragraph = \"\"\"I have three visions for India. In 3000 years of our history, people from all over \n the world have come and invaded us, captured our lands, conquered our minds. \n From Alexander onwards, the Greeks, the Turks, the Moguls, the Portuguese, the British,\n the French, the Dutch, all of them came and looted us, took over what was ours. \n Yet we have not done this to any other nation. We have not conquered anyone. \n We have not grabbed their land, their culture, \n their history and tried to enforce our way of life on them. \n Why? Because we respect the freedom of others.That is why my \n first vision is that of freedom. I believe that India got its first vision of \n this in 1857, when we started the War of Independence. It is this freedom that\n we must protect and nurture and build on. If we are not free, no one will respect us.\n My second vision for India’s development. For fifty years we have been a developing nation.\n It is time we see ourselves as a developed nation. We are among the top 5 nations of the world\n in terms of GDP. We have a 10 percent growth rate in most areas. Our poverty levels are falling.\n Our achievements are being globally recognised today. Yet we lack the self-confidence to\n see ourselves as a developed nation, self-reliant and self-assured. Isn’t this incorrect?\n I have a third vision. India must stand up to the world. Because I believe that unless India \n stands up to the world, no one will respect us. Only strength respects strength. We must be \n strong not only as a military power but also as an economic power. Both must go hand-in-hand. \n My good fortune was to have worked with three great minds. Dr. Vikram Sarabhai of the Dept. of \n space, Professor Satish Dhawan, who succeeded him and Dr. Brahm Prakash, father of nuclear material.\n I was lucky to have worked with all three of them closely and consider this the great opportunity of my life. \n I see four milestones in my career\"\"\"",
"_____no_output_____"
],
[
"sentences=nltk.sent_tokenize(paragraph)",
"_____no_output_____"
],
[
"ps=PorterStemmer()",
"_____no_output_____"
],
[
"for i in range(len(sentences)):\n words=nltk.word_tokenize(paragraph)\n words=[ps.stem(word) for word in words if not word in set(stopwords.words('english'))]\n sentences=' '.join(words)",
"_____no_output_____"
],
[
"sentences",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d00076bca8d2b781f0ba8adff988c49a32fc6928 | 9,146 | ipynb | Jupyter Notebook | jupyter/onnxruntime/machine_learning_with_ONNXRuntime.ipynb | raghav-deepsource/djl | 8d774578a51b298d2ddeb1a898ddd5a157b7f0bd | [
"Apache-2.0"
] | 1 | 2020-11-25T06:01:52.000Z | 2020-11-25T06:01:52.000Z | jupyter/onnxruntime/machine_learning_with_ONNXRuntime.ipynb | wulin-challenge/djl | 5dd343ccc03a75322efcd441b6f5234339bd95f3 | [
"Apache-2.0"
] | null | null | null | jupyter/onnxruntime/machine_learning_with_ONNXRuntime.ipynb | wulin-challenge/djl | 5dd343ccc03a75322efcd441b6f5234339bd95f3 | [
"Apache-2.0"
] | null | null | null | 39.765217 | 439 | 0.622677 | [
[
[
"# Classification on Iris dataset with sklearn and DJL\n\nIn this notebook, you will try to use a pre-trained sklearn model to run on DJL for a general classification task. The model was trained with [Iris flower dataset](https://en.wikipedia.org/wiki/Iris_flower_data_set).\n\n## Background \n\n### Iris Dataset\n\nThe dataset contains a set of 150 records under five attributes - sepal length, sepal width, petal length, petal width and species.\n\nIris setosa | Iris versicolor | Iris virginica\n:-------------------------:|:-------------------------:|:-------------------------:\n |  |  \n\nThe chart above shows three different kinds of the Iris flowers. \n\nWe will use sepal length, sepal width, petal length, petal width as the feature and species as the label to train the model.\n\n### Sklearn Model\n\nYou can find more information [here](http://onnx.ai/sklearn-onnx/). You can use the sklearn built-in iris dataset to load the data. Then we defined a [RandomForestClassifer](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html) to train the model. After that, we convert the model to onnx format for DJL to run inference. The following code is a sample classification setup using sklearn:\n\n```python\n# Train a model.\nfrom sklearn.datasets import load_iris\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.ensemble import RandomForestClassifier\niris = load_iris()\nX, y = iris.data, iris.target\nX_train, X_test, y_train, y_test = train_test_split(X, y)\nclr = RandomForestClassifier()\nclr.fit(X_train, y_train)\n```\n\n\n## Preparation\n\nThis tutorial requires the installation of Java Kernel. To install the Java Kernel, see the [README](https://github.com/awslabs/djl/blob/master/jupyter/README.md).\n\nThese are dependencies we will use. To enhance the NDArray operation capability, we are importing ONNX Runtime and PyTorch Engine at the same time. Please find more information [here](https://github.com/awslabs/djl/blob/master/docs/onnxruntime/hybrid_engine.md#hybrid-engine-for-onnx-runtime).",
"_____no_output_____"
]
],
[
[
"// %mavenRepo snapshots https://oss.sonatype.org/content/repositories/snapshots/\n\n%maven ai.djl:api:0.8.0\n%maven ai.djl.onnxruntime:onnxruntime-engine:0.8.0\n%maven ai.djl.pytorch:pytorch-engine:0.8.0\n%maven org.slf4j:slf4j-api:1.7.26\n%maven org.slf4j:slf4j-simple:1.7.26\n\n%maven com.microsoft.onnxruntime:onnxruntime:1.4.0\n%maven ai.djl.pytorch:pytorch-native-auto:1.6.0",
"_____no_output_____"
],
[
"import ai.djl.inference.*;\nimport ai.djl.modality.*;\nimport ai.djl.ndarray.*;\nimport ai.djl.ndarray.types.*;\nimport ai.djl.repository.zoo.*;\nimport ai.djl.translate.*;\nimport java.util.*;",
"_____no_output_____"
]
],
[
[
"## Step 1 create a Translator\n\nInference in machine learning is the process of predicting the output for a given input based on a pre-defined model.\nDJL abstracts away the whole process for ease of use. It can load the model, perform inference on the input, and provide\noutput. DJL also allows you to provide user-defined inputs. The workflow looks like the following:\n\n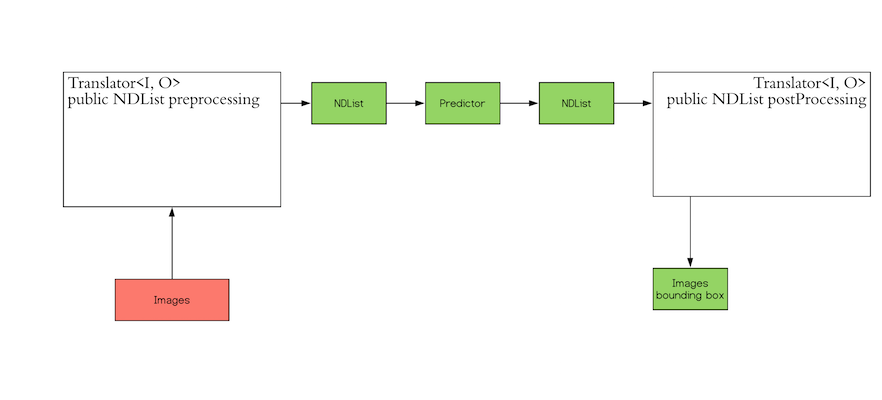\n\nThe `Translator` interface encompasses the two white blocks: Pre-processing and Post-processing. The pre-processing\ncomponent converts the user-defined input objects into an NDList, so that the `Predictor` in DJL can understand the\ninput and make its prediction. Similarly, the post-processing block receives an NDList as the output from the\n`Predictor`. The post-processing block allows you to convert the output from the `Predictor` to the desired output\nformat.\n\nIn our use case, we use a class namely `IrisFlower` as our input class type. We will use [`Classifications`](https://javadoc.io/doc/ai.djl/api/latest/ai/djl/modality/Classifications.html) as our output class type.",
"_____no_output_____"
]
],
[
[
"public static class IrisFlower {\n\n public float sepalLength;\n public float sepalWidth;\n public float petalLength;\n public float petalWidth;\n\n public IrisFlower(float sepalLength, float sepalWidth, float petalLength, float petalWidth) {\n this.sepalLength = sepalLength;\n this.sepalWidth = sepalWidth;\n this.petalLength = petalLength;\n this.petalWidth = petalWidth;\n }\n}",
"_____no_output_____"
]
],
[
[
"Let's create a translator",
"_____no_output_____"
]
],
[
[
"public static class MyTranslator implements Translator<IrisFlower, Classifications> {\n\n private final List<String> synset;\n\n public MyTranslator() {\n // species name\n synset = Arrays.asList(\"setosa\", \"versicolor\", \"virginica\");\n }\n\n @Override\n public NDList processInput(TranslatorContext ctx, IrisFlower input) {\n float[] data = {input.sepalLength, input.sepalWidth, input.petalLength, input.petalWidth};\n NDArray array = ctx.getNDManager().create(data, new Shape(1, 4));\n return new NDList(array);\n }\n\n @Override\n public Classifications processOutput(TranslatorContext ctx, NDList list) {\n return new Classifications(synset, list.get(1));\n }\n\n @Override\n public Batchifier getBatchifier() {\n return null;\n }\n}",
"_____no_output_____"
]
],
[
[
"## Step 2 Prepare your model\n\nWe will load a pretrained sklearn model into DJL. We defined a [`ModelZoo`](https://javadoc.io/doc/ai.djl/api/latest/ai/djl/repository/zoo/ModelZoo.html) concept to allow user load model from varity of locations, such as remote URL, local files or DJL pretrained model zoo. We need to define `Criteria` class to help the modelzoo locate the model and attach translator. In this example, we download a compressed ONNX model from S3.",
"_____no_output_____"
]
],
[
[
"String modelUrl = \"https://mlrepo.djl.ai/model/tabular/random_forest/ai/djl/onnxruntime/iris_flowers/0.0.1/iris_flowers.zip\";\nCriteria<IrisFlower, Classifications> criteria = Criteria.builder()\n .setTypes(IrisFlower.class, Classifications.class)\n .optModelUrls(modelUrl)\n .optTranslator(new MyTranslator())\n .optEngine(\"OnnxRuntime\") // use OnnxRuntime engine by default\n .build();\nZooModel<IrisFlower, Classifications> model = ModelZoo.loadModel(criteria);",
"_____no_output_____"
]
],
[
[
"## Step 3 Run inference\n\nUser will just need to create a `Predictor` from model to run the inference.",
"_____no_output_____"
]
],
[
[
"Predictor<IrisFlower, Classifications> predictor = model.newPredictor();\nIrisFlower info = new IrisFlower(1.0f, 2.0f, 3.0f, 4.0f);\npredictor.predict(info);",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d00080cae9b7a28ebc8ef5ae33eb9e79b8f215bf | 5,019 | ipynb | Jupyter Notebook | Algorithms/landsat_radiance.ipynb | OIEIEIO/earthengine-py-notebooks | 5d6c5cdec0c73bf02020ee17d42c9e30d633349f | [
"MIT"
] | 1,008 | 2020-01-27T02:03:18.000Z | 2022-03-24T10:42:14.000Z | Algorithms/landsat_radiance.ipynb | rafatieppo/earthengine-py-notebooks | 99fbc4abd1fb6ba41e3d8a55f8911217353a3237 | [
"MIT"
] | 8 | 2020-02-01T20:18:18.000Z | 2021-11-23T01:48:02.000Z | Algorithms/landsat_radiance.ipynb | rafatieppo/earthengine-py-notebooks | 99fbc4abd1fb6ba41e3d8a55f8911217353a3237 | [
"MIT"
] | 325 | 2020-01-27T02:03:36.000Z | 2022-03-25T20:33:33.000Z | 36.635036 | 470 | 0.557282 | [
[
[
"<table class=\"ee-notebook-buttons\" align=\"left\">\n <td><a target=\"_blank\" href=\"https://github.com/giswqs/earthengine-py-notebooks/tree/master/Algorithms/landsat_radiance.ipynb\"><img width=32px src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" /> View source on GitHub</a></td>\n <td><a target=\"_blank\" href=\"https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Algorithms/landsat_radiance.ipynb\"><img width=26px src=\"https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png\" />Notebook Viewer</a></td>\n <td><a target=\"_blank\" href=\"https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Algorithms/landsat_radiance.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" /> Run in Google Colab</a></td>\n</table>",
"_____no_output_____"
],
[
"## Install Earth Engine API and geemap\nInstall the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://geemap.org). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.\nThe following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.",
"_____no_output_____"
]
],
[
[
"# Installs geemap package\nimport subprocess\n\ntry:\n import geemap\nexcept ImportError:\n print('Installing geemap ...')\n subprocess.check_call([\"python\", '-m', 'pip', 'install', 'geemap'])",
"_____no_output_____"
],
[
"import ee\nimport geemap",
"_____no_output_____"
]
],
[
[
"## Create an interactive map \nThe default basemap is `Google Maps`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/basemaps.py) can be added using the `Map.add_basemap()` function. ",
"_____no_output_____"
]
],
[
[
"Map = geemap.Map(center=[40,-100], zoom=4)\nMap",
"_____no_output_____"
]
],
[
[
"## Add Earth Engine Python script ",
"_____no_output_____"
]
],
[
[
"# Add Earth Engine dataset\n# Load a raw Landsat scene and display it.\nraw = ee.Image('LANDSAT/LC08/C01/T1/LC08_044034_20140318')\nMap.centerObject(raw, 10)\nMap.addLayer(raw, {'bands': ['B4', 'B3', 'B2'], 'min': 6000, 'max': 12000}, 'raw')\n\n# Convert the raw data to radiance.\nradiance = ee.Algorithms.Landsat.calibratedRadiance(raw)\nMap.addLayer(radiance, {'bands': ['B4', 'B3', 'B2'], 'max': 90}, 'radiance')\n\n# Convert the raw data to top-of-atmosphere reflectance.\ntoa = ee.Algorithms.Landsat.TOA(raw)\n\nMap.addLayer(toa, {'bands': ['B4', 'B3', 'B2'], 'max': 0.2}, 'toa reflectance')\n\n",
"_____no_output_____"
]
],
[
[
"## Display Earth Engine data layers ",
"_____no_output_____"
]
],
[
[
"Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.\nMap",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0008b5894090e9887e8ce1ff35481414c1bb8d4 | 22,698 | ipynb | Jupyter Notebook | cp2/cp2_method0.ipynb | jet-code/multivariable-control-systems | 81b57d51a4dfc92964f989794f71d525af0359ff | [
"MIT"
] | null | null | null | cp2/cp2_method0.ipynb | jet-code/multivariable-control-systems | 81b57d51a4dfc92964f989794f71d525af0359ff | [
"MIT"
] | null | null | null | cp2/cp2_method0.ipynb | jet-code/multivariable-control-systems | 81b57d51a4dfc92964f989794f71d525af0359ff | [
"MIT"
] | null | null | null | 22.858006 | 89 | 0.390475 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0009800054b678bfad6c1462b810393ddac51b0 | 217,601 | ipynb | Jupyter Notebook | MNIST/Session2/3_Global_Average_Pooling.ipynb | gmshashank/pytorch_vision | 54367b83e9780fe14c6f8b93157091ffdf7266eb | [
"MIT"
] | null | null | null | MNIST/Session2/3_Global_Average_Pooling.ipynb | gmshashank/pytorch_vision | 54367b83e9780fe14c6f8b93157091ffdf7266eb | [
"MIT"
] | null | null | null | MNIST/Session2/3_Global_Average_Pooling.ipynb | gmshashank/pytorch_vision | 54367b83e9780fe14c6f8b93157091ffdf7266eb | [
"MIT"
] | null | null | null | 101.209767 | 53,662 | 0.792552 | [
[
[
"# Import Libraries",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nimport torchvision\nfrom torchvision import datasets, transforms",
"_____no_output_____"
],
[
"%matplotlib inline\r\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Data Transformations\n\nWe first start with defining our data transformations. We need to think what our data is and how can we augment it to correct represent images which it might not see otherwise. \n",
"_____no_output_____"
]
],
[
[
"# Train Phase transformations\ntrain_transforms = transforms.Compose([\n # transforms.Resize((28, 28)),\n # transforms.ColorJitter(brightness=0.10, contrast=0.1, saturation=0.10, hue=0.1),\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,)) # The mean and std have to be sequences (e.g., tuples), therefore you should add a comma after the values. \n # Note the difference between (0.1307) and (0.1307,)\n ])\n\n# Test Phase transformations\ntest_transforms = transforms.Compose([\n # transforms.Resize((28, 28)),\n # transforms.ColorJitter(brightness=0.10, contrast=0.1, saturation=0.10, hue=0.1),\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,))\n ])\n",
"_____no_output_____"
]
],
[
[
"# Dataset and Creating Train/Test Split",
"_____no_output_____"
]
],
[
[
"train = datasets.MNIST('./data', train=True, download=True, transform=train_transforms)\ntest = datasets.MNIST('./data', train=False, download=True, transform=test_transforms)",
"Downloading http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz to ./data/MNIST/raw/train-images-idx3-ubyte.gz\n"
]
],
[
[
"# Dataloader Arguments & Test/Train Dataloaders\n",
"_____no_output_____"
]
],
[
[
"SEED = 1\n\n# CUDA?\ncuda = torch.cuda.is_available()\nprint(\"CUDA Available?\", cuda)\n\n# For reproducibility\ntorch.manual_seed(SEED)\n\nif cuda:\n torch.cuda.manual_seed(SEED)\n\n# dataloader arguments - something you'll fetch these from cmdprmt\ndataloader_args = dict(shuffle=True, batch_size=128, num_workers=4, pin_memory=True) if cuda else dict(shuffle=True, batch_size=64)\n\n# train dataloader\ntrain_loader = torch.utils.data.DataLoader(train, **dataloader_args)\n\n# test dataloader\ntest_loader = torch.utils.data.DataLoader(test, **dataloader_args)",
"CUDA Available? True\n"
]
],
[
[
"# Data Statistics\n\nIt is important to know your data very well. Let's check some of the statistics around our data and how it actually looks like",
"_____no_output_____"
]
],
[
[
"# We'd need to convert it into Numpy! Remember above we have converted it into tensors already\ntrain_data = train.train_data\ntrain_data = train.transform(train_data.numpy())\n\nprint('[Train]')\nprint(' - Numpy Shape:', train.train_data.cpu().numpy().shape)\nprint(' - Tensor Shape:', train.train_data.size())\nprint(' - min:', torch.min(train_data))\nprint(' - max:', torch.max(train_data))\nprint(' - mean:', torch.mean(train_data))\nprint(' - std:', torch.std(train_data))\nprint(' - var:', torch.var(train_data))\n\ndataiter = iter(train_loader)\nimages, labels = dataiter.next()\n\nprint(images.shape)\nprint(labels.shape)\n\n# Let's visualize some of the images\nplt.imshow(images[0].numpy().squeeze(), cmap='gray_r')",
"\n"
]
],
[
[
"## MORE\n\nIt is important that we view as many images as possible. This is required to get some idea on image augmentation later on",
"_____no_output_____"
]
],
[
[
"figure = plt.figure()\nnum_of_images = 60\nfor index in range(1, num_of_images + 1):\n plt.subplot(6, 10, index)\n plt.axis('off')\n plt.imshow(images[index].numpy().squeeze(), cmap='gray_r')",
"_____no_output_____"
]
],
[
[
"# The model\nLet's start with the model we first saw",
"_____no_output_____"
]
],
[
[
"class Net(nn.Module):\n def __init__(self):\n super(Net, self).__init__()\n # Input Block\n self.convblock1 = nn.Sequential(\n nn.Conv2d(in_channels=1, out_channels=16, kernel_size=(3, 3), padding=0, bias=False),\n nn.ReLU(),\n ) # output_size = 26\n\n # CONVOLUTION BLOCK 1\n self.convblock2 = nn.Sequential(\n nn.Conv2d(in_channels=16, out_channels=16, kernel_size=(3, 3), padding=0, bias=False),\n nn.ReLU(),\n ) # output_size = 24\n\n # TRANSITION BLOCK 1\n self.convblock3 = nn.Sequential(\n nn.Conv2d(in_channels=16, out_channels=16, kernel_size=(1, 1), padding=0, bias=False),\n nn.ReLU(),\n ) # output_size = 24\n self.pool1 = nn.MaxPool2d(2, 2) # output_size = 12\n\n # CONVOLUTION BLOCK 2\n self.convblock4 = nn.Sequential(\n nn.Conv2d(in_channels=16, out_channels=16, kernel_size=(3, 3), padding=0, bias=False),\n nn.ReLU(),\n ) # output_size = 10\n\n self.convblock5 = nn.Sequential(\n nn.Conv2d(in_channels=16, out_channels=16, kernel_size=(3, 3), padding=0, bias=False),\n nn.ReLU(),\n ) # output_size = 8\n self.convblock6 = nn.Sequential(\n nn.Conv2d(in_channels=16, out_channels=10, kernel_size=(3, 3), padding=0, bias=False),\n nn.ReLU(),\n ) # output_size = 6\n\n # OUTPUT BLOCK\n self.convblock7 = nn.Sequential(\n nn.Conv2d(in_channels=10, out_channels=10, kernel_size=(3, 3), padding=1, bias=False),\n nn.ReLU(),\n ) # output_size = 6\n\n self.gap = nn.Sequential(\n nn.AvgPool2d(kernel_size=6)\n )\n\n self.convblock8 = nn.Sequential(\n nn.Conv2d(in_channels=10, out_channels=10, kernel_size=(1, 1), padding=0, bias=False),\n # nn.BatchNorm2d(10), NEVER\n # nn.ReLU() NEVER!\n ) # output_size = 1\n\n def forward(self, x):\n x = self.convblock1(x)\n x = self.convblock2(x)\n x = self.convblock3(x)\n x = self.pool1(x)\n x = self.convblock4(x)\n x = self.convblock5(x)\n x = self.convblock6(x)\n x = self.convblock7(x)\n x = self.gap(x)\n x = self.convblock8(x)\n x = x.view(-1, 10)\n return F.log_softmax(x, dim=-1)",
"_____no_output_____"
]
],
[
[
"# Model Params\nCan't emphasize on how important viewing Model Summary is. \nUnfortunately, there is no in-built model visualizer, so we have to take external help",
"_____no_output_____"
]
],
[
[
"!pip install torchsummary\nfrom torchsummary import summary\nuse_cuda = torch.cuda.is_available()\ndevice = torch.device(\"cuda\" if use_cuda else \"cpu\")\nprint(device)\nmodel = Net().to(device)\nsummary(model, input_size=(1, 28, 28))\n",
"Requirement already satisfied: torchsummary in /usr/local/lib/python3.6/dist-packages (1.5.1)\ncuda\n----------------------------------------------------------------\n Layer (type) Output Shape Param #\n================================================================\n Conv2d-1 [-1, 16, 26, 26] 144\n ReLU-2 [-1, 16, 26, 26] 0\n Conv2d-3 [-1, 16, 24, 24] 2,304\n ReLU-4 [-1, 16, 24, 24] 0\n Conv2d-5 [-1, 16, 24, 24] 256\n ReLU-6 [-1, 16, 24, 24] 0\n MaxPool2d-7 [-1, 16, 12, 12] 0\n Conv2d-8 [-1, 16, 10, 10] 2,304\n ReLU-9 [-1, 16, 10, 10] 0\n Conv2d-10 [-1, 16, 8, 8] 2,304\n ReLU-11 [-1, 16, 8, 8] 0\n Conv2d-12 [-1, 10, 6, 6] 1,440\n ReLU-13 [-1, 10, 6, 6] 0\n Conv2d-14 [-1, 10, 6, 6] 900\n ReLU-15 [-1, 10, 6, 6] 0\n AvgPool2d-16 [-1, 10, 1, 1] 0\n Conv2d-17 [-1, 10, 1, 1] 100\n================================================================\nTotal params: 9,752\nTrainable params: 9,752\nNon-trainable params: 0\n----------------------------------------------------------------\nInput size (MB): 0.00\nForward/backward pass size (MB): 0.52\nParams size (MB): 0.04\nEstimated Total Size (MB): 0.56\n----------------------------------------------------------------\n"
]
],
[
[
"# Training and Testing\n\nLooking at logs can be boring, so we'll introduce **tqdm** progressbar to get cooler logs. \n\nLet's write train and test functions",
"_____no_output_____"
]
],
[
[
"from tqdm import tqdm\n\ntrain_losses = []\ntest_losses = []\ntrain_acc = []\ntest_acc = []\n\ndef train(model, device, train_loader, optimizer, epoch):\n\n global train_max\n model.train()\n pbar = tqdm(train_loader)\n correct = 0\n processed = 0\n for batch_idx, (data, target) in enumerate(pbar):\n # get samples\n data, target = data.to(device), target.to(device)\n\n # Init\n optimizer.zero_grad()\n # In PyTorch, we need to set the gradients to zero before starting to do backpropragation because PyTorch accumulates the gradients on subsequent backward passes. \n # Because of this, when you start your training loop, ideally you should zero out the gradients so that you do the parameter update correctly.\n\n # Predict\n y_pred = model(data)\n\n # Calculate loss\n loss = F.nll_loss(y_pred, target)\n train_losses.append(loss)\n \n # Backpropagation\n loss.backward()\n optimizer.step()\n\n # Update pbar-tqdm\n \n pred = y_pred.argmax(dim=1, keepdim=True) # get the index of the max log-probability\n correct += pred.eq(target.view_as(pred)).sum().item()\n processed += len(data)\n \n pbar.set_description(desc= f'Loss={loss.item()} Batch_id={batch_idx} Accuracy={100*correct/processed:0.2f}')\n train_acc.append(100*correct/processed)\n \n if (train_max < 100*correct/processed):\n train_max = 100*correct/processed\n\n\ndef test(model, device, test_loader):\n\n global test_max\n model.eval()\n test_loss = 0\n correct = 0\n with torch.no_grad():\n for data, target in test_loader:\n data, target = data.to(device), target.to(device)\n output = model(data)\n test_loss += F.nll_loss(output, target, reduction='sum').item() # sum up batch loss\n pred = output.argmax(dim=1, keepdim=True) # get the index of the max log-probability\n correct += pred.eq(target.view_as(pred)).sum().item()\n\n test_loss /= len(test_loader.dataset)\n test_losses.append(test_loss)\n\n print('\\nTest set: Average loss: {:.4f}, Accuracy: {}/{} ({:.2f}%)\\n'.format(\n test_loss, correct, len(test_loader.dataset),\n 100. * correct / len(test_loader.dataset)))\n\n if (test_max < 100. * correct / len(test_loader.dataset)):\n test_max = 100. * correct / len(test_loader.dataset)\n \n test_acc.append(100. * correct / len(test_loader.dataset))\n",
"_____no_output_____"
]
],
[
[
"# Let's Train and test our model",
"_____no_output_____"
]
],
[
[
"model = Net().to(device)\noptimizer = optim.SGD(model.parameters(), lr=0.01, momentum=0.9)\nEPOCHS = 15\ntrain_max=0\ntest_max=0\nfor epoch in range(EPOCHS):\n print(\"EPOCH:\", epoch)\n train(model, device, train_loader, optimizer, epoch)\n test(model, device, test_loader)\n\nprint(f\"\\nMaximum training accuracy: {train_max}\\n\")\nprint(f\"\\nMaximum test accuracy: {test_max}\\n\")\n",
"\r 0%| | 0/469 [00:00<?, ?it/s]"
],
[
"fig, axs = plt.subplots(2,2,figsize=(15,10))\naxs[0, 0].plot(train_losses)\naxs[0, 0].set_title(\"Training Loss\")\naxs[1, 0].plot(train_acc)\naxs[1, 0].set_title(\"Training Accuracy\")\naxs[0, 1].plot(test_losses)\naxs[0, 1].set_title(\"Test Loss\")\naxs[1, 1].plot(test_acc)\naxs[1, 1].set_title(\"Test Accuracy\")",
"_____no_output_____"
],
[
"fig, ((axs1, axs2), (axs3, axs4)) = plt.subplots(2,2,figsize=(15,10)) \r\n# Train plot\r\naxs1.plot(train_losses)\r\naxs1.set_title(\"Training Loss\")\r\naxs3.plot(train_acc)\r\naxs3.set_title(\"Training Accuracy\")\r\n\r\n# axs1.set_xlim([0, 5])\r\naxs1.set_ylim([0, 5])\r\naxs3.set_ylim([0, 100])\r\n\r\n\r\n# Test plot\r\naxs2.plot(test_losses)\r\naxs2.set_title(\"Test Loss\")\r\naxs4.plot(test_acc)\r\naxs4.set_title(\"Test Accuracy\")\r\n\r\naxs2.set_ylim([0, 5])\r\naxs4.set_ylim([0, 100])\r\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0009c796f5114ea1df1abddfe294ea1d19ec655 | 202,823 | ipynb | Jupyter Notebook | src/archive/CAE.ipynb | RiceD2KLab/TCH_CardiacSignals_F20 | ea6e84703086ddb7bfc5ba164aa67acdc9e78b7d | [
"BSD-2-Clause"
] | 1 | 2022-01-27T07:03:20.000Z | 2022-01-27T07:03:20.000Z | src/archive/CAE.ipynb | RiceD2KLab/TCH_CardiacSignals_F20 | ea6e84703086ddb7bfc5ba164aa67acdc9e78b7d | [
"BSD-2-Clause"
] | null | null | null | src/archive/CAE.ipynb | RiceD2KLab/TCH_CardiacSignals_F20 | ea6e84703086ddb7bfc5ba164aa67acdc9e78b7d | [
"BSD-2-Clause"
] | null | null | null | 270.791722 | 19,223 | 0.600134 | [
[
[
"%cd /Users/Kunal/Projects/TCH_CardiacSignals_F20/",
"/Users/kunal/Projects/TCH_CardiacSignals_F20\n"
],
[
"from numpy.random import seed\nseed(1)\nimport numpy as np\nimport os\nimport matplotlib.pyplot as plt\nimport tensorflow\ntensorflow.random.set_seed(2)\nfrom tensorflow import keras\nfrom tensorflow.keras.callbacks import EarlyStopping\nfrom tensorflow.keras.regularizers import l1, l2\nfrom tensorflow.keras.layers import Dense, Flatten, Reshape, Input, InputLayer, Dropout, Conv1D, MaxPooling1D, BatchNormalization, UpSampling1D, Conv1DTranspose\nfrom tensorflow.keras.models import Sequential, Model\nfrom src.preprocess.dim_reduce.patient_split import *\nfrom src.preprocess.heartbeat_split import heartbeat_split\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"data = np.load(\"Working_Data/Training_Subset/Normalized/ten_hbs/Normalized_Fixed_Dim_HBs_Idx\" + str(1) + \".npy\")\ndata.shape",
"_____no_output_____"
],
[
"def read_in(file_index, normalized, train, ratio):\n \"\"\"\n Reads in a file and can toggle between normalized and original files\n :param file_index: patient number as string\n :param normalized: binary that determines whether the files should be normalized or not\n :param train: int that determines whether or not we are reading in data to train the model or for encoding\n :param ratio: ratio to split the files into train and test\n :return: returns npy array of patient data across 4 leads\n \"\"\"\n # filepath = os.path.join(\"Working_Data\", \"Normalized_Fixed_Dim_HBs_Idx\" + file_index + \".npy\")\n # filepath = os.path.join(\"Working_Data\", \"1000d\", \"Normalized_Fixed_Dim_HBs_Idx35.npy\")\n filepath = \"Working_Data/Training_Subset/Normalized/ten_hbs/Normalized_Fixed_Dim_HBs_Idx\" + str(file_index) + \".npy\"\n\n if normalized == 1:\n if train == 1:\n normal_train, normal_test, abnormal = patient_split_train(filepath, ratio)\n # noise_factor = 0.5\n # noise_train = normal_train + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=normal_train.shape)\n return normal_train, normal_test\n elif train == 0:\n training, test, full = patient_split_all(filepath, ratio)\n return training, test, full\n elif train == 2:\n train_, test, full = patient_split_all(filepath, ratio)\n # 4x the data\n noise_factor = 0.5\n\n noise_train = train_ + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=train_.shape)\n noise_train2 = train_ + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=train_.shape)\n noise_train3 = train_ + noise_factor * np.random.normal(loc=0.0, scale=1.0, size=train_.shape)\n train_ = np.concatenate((train_, noise_train, noise_train2, noise_train3))\n return train_, test, full\n else:\n data = np.load(os.path.join(\"Working_Data\", \"Fixed_Dim_HBs_Idx\" + file_index + \".npy\"))\n return data",
"_____no_output_____"
],
[
"\ndef build_model(sig_shape, encode_size):\n \"\"\"\n Builds a deterministic autoencoder model, returning both the encoder and decoder models\n :param sig_shape: shape of input signal\n :param encode_size: dimension that we want to reduce to\n :return: encoder, decoder models\n \"\"\"\n # encoder = Sequential()\n # encoder.add(InputLayer((1000,4)))\n # # idk if causal is really making that much of an impact but it seems useful for time series data?\n # encoder.add(Conv1D(10, 11, activation=\"linear\", padding=\"causal\"))\n # encoder.add(Conv1D(10, 5, activation=\"relu\", padding=\"causal\"))\n # # encoder.add(Conv1D(10, 3, activation=\"relu\", padding=\"same\"))\n # encoder.add(Flatten())\n # encoder.add(Dense(750, activation = 'tanh', kernel_initializer='glorot_normal')) #tanh\n # encoder.add(Dense(500, activation='relu', kernel_initializer='glorot_normal'))\n # encoder.add(Dense(400, activation = 'relu', kernel_initializer='glorot_normal'))\n # encoder.add(Dense(300, activation='relu', kernel_initializer='glorot_normal'))\n # encoder.add(Dense(200, activation = 'relu', kernel_initializer='glorot_normal')) #relu\n # encoder.add(Dense(encode_size))\n\n\n encoder = Sequential()\n encoder.add(InputLayer((1000,4)))\n encoder.add(Conv1D(3, 11, activation=\"tanh\", padding=\"same\"))\n encoder.add(Conv1D(5, 7, activation=\"relu\", padding=\"same\"))\n encoder.add(MaxPooling1D(2))\n encoder.add(Conv1D(5, 5, activation=\"tanh\", padding=\"same\"))\n encoder.add(Conv1D(7, 3, activation=\"tanh\", padding=\"same\"))\n encoder.add(MaxPooling1D(2))\n encoder.add(Flatten())\n encoder.add(Dense(750, activation = 'tanh', kernel_initializer='glorot_normal'))\n # encoder.add(Dense(500, activation='relu', kernel_initializer='glorot_normal'))\n encoder.add(Dense(400, activation = 'tanh', kernel_initializer='glorot_normal'))\n # encoder.add(Dense(300, activation='relu', kernel_initializer='glorot_normal'))\n encoder.add(Dense(200, activation = 'tanh', kernel_initializer='glorot_normal'))\n encoder.add(Dense(encode_size))\n # encoder.summary()\n ####################################################################################################################\n # Build the decoder\n\n # decoder = Sequential()\n # decoder.add(InputLayer((latent_dim,)))\n # decoder.add(Dense(200, activation='tanh', kernel_initializer='glorot_normal'))\n # decoder.add(Dense(300, activation='relu', kernel_initializer='glorot_normal'))\n # decoder.add(Dense(400, activation='relu', kernel_initializer='glorot_normal'))\n # decoder.add(Dense(500, activation='relu', kernel_initializer='glorot_normal'))\n # decoder.add(Dense(750, activation='relu', kernel_initializer='glorot_normal'))\n # decoder.add(Dense(10000, activation='relu', kernel_initializer='glorot_normal'))\n # decoder.add(Reshape((1000, 10)))\n # decoder.add(Conv1DTranspose(4, 7, activation=\"relu\", padding=\"same\"))\n\n decoder = Sequential()\n decoder.add(InputLayer((encode_size,)))\n decoder.add(Dense(200, activation='tanh', kernel_initializer='glorot_normal'))\n # decoder.add(Dense(300, activation='relu', kernel_initializer='glorot_normal'))\n decoder.add(Dense(400, activation='tanh', kernel_initializer='glorot_normal'))\n # decoder.add(Dense(500, activation='relu', kernel_initializer='glorot_normal'))\n decoder.add(Dense(750, activation='tanh', kernel_initializer='glorot_normal'))\n decoder.add(Dense(10000, activation='tanh', kernel_initializer='glorot_normal'))\n decoder.add(Reshape((1000, 10)))\n # decoder.add(Conv1DTranspose(8, 3, activation=\"relu\", padding=\"same\"))\n decoder.add(Conv1DTranspose(8, 11, activation=\"relu\", padding=\"same\"))\n decoder.add(Conv1DTranspose(4, 5, activation=\"linear\", padding=\"same\"))\n\n return encoder, decoder",
"_____no_output_____"
],
[
"def training_ae(num_epochs, reduced_dim, file_index):\n \"\"\"\n Training function for deterministic autoencoder model, saves the encoded and reconstructed arrays\n :param num_epochs: number of epochs to use\n :param reduced_dim: goal dimension\n :param file_index: patient number\n :return: None\n \"\"\"\n normal, abnormal, all = read_in(file_index, 1, 2, 0.3)\n normal_train, normal_valid = train_test_split(normal, train_size=0.85, random_state=1)\n # normal_train = normal[:round(len(normal)*.85),:]\n # normal_valid = normal[round(len(normal)*.85):,:]\n signal_shape = normal.shape[1:]\n batch_size = round(len(normal) * 0.1)\n\n encoder, decoder = build_model(signal_shape, reduced_dim)\n\n inp = Input(signal_shape)\n encode = encoder(inp)\n reconstruction = decoder(encode)\n\n autoencoder = Model(inp, reconstruction)\n opt = keras.optimizers.Adam(learning_rate=0.0001) #0.0008\n autoencoder.compile(optimizer=opt, loss='mse')\n\n early_stopping = EarlyStopping(patience=10, min_delta=0.001, mode='min')\n autoencoder = autoencoder.fit(x=normal_train, y=normal_train, epochs=num_epochs, validation_data=(normal_valid, normal_valid), batch_size=batch_size, callbacks=early_stopping)\n\n plt.plot(autoencoder.history['loss'])\n plt.plot(autoencoder.history['val_loss'])\n plt.title('model loss patient' + str(file_index))\n plt.ylabel('loss')\n plt.xlabel('epoch')\n plt.legend(['train', 'validation'], loc='upper left')\n plt.show()\n\n # using AE to encode other data\n encoded = encoder.predict(all)\n reconstruction = decoder.predict(encoded)\n\n # save reconstruction, encoded, and input if needed\n # reconstruction_save = os.path.join(\"Working_Data\", \"reconstructed_ae_\" + str(reduced_dim) + \"d_Idx\" + str(file_index) + \".npy\")\n # encoded_save = os.path.join(\"Working_Data\", \"reduced_ae_\" + str(reduced_dim) + \"d_Idx\" + str(file_index) + \".npy\")\n\n reconstruction_save = \"Working_Data/Training_Subset/Model_Output/reconstructed_10hb_cae_\" + str(file_index) + \".npy\"\n encoded_save = \"Working_Data/Training_Subset/Model_Output/encoded_10hb_cae_\" + str(file_index) + \".npy\"\n\n np.save(reconstruction_save, reconstruction)\n np.save(encoded_save,encoded)\n\n # if training and need to save test split for MSE calculation\n # input_save = os.path.join(\"Working_Data\",\"1000d\", \"original_data_test_ae\" + str(100) + \"d_Idx\" + str(35) + \".npy\")\n # np.save(input_save, test)",
"_____no_output_____"
],
[
"def run(num_epochs, encoded_dim):\n \"\"\"\n Run training autoencoder over all dims in list\n :param num_epochs: number of epochs to train for\n :param encoded_dim: dimension to run on\n :return None, saves arrays for reconstructed and dim reduced arrays\n \"\"\"\n for patient_ in [1,16,4,11]: #heartbeat_split.indicies:\n print(\"Starting on index: \" + str(patient_))\n training_ae(num_epochs, encoded_dim, patient_)\n print(\"Completed \" + str(patient_) + \" reconstruction and encoding, saved test data to assess performance\")\n\n",
"_____no_output_____"
],
[
"#################### Training to be done for 100 epochs for all dimensions ############################################\nrun(100, 100)\n\n# run(100,100)",
"Starting on index: 1\nEpoch 1/100\n9/9 [==============================] - 13s 1s/step - loss: 1.1561 - val_loss: 1.0856\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 2/100\n9/9 [==============================] - 9s 949ms/step - loss: 1.0506 - val_loss: 1.0042\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 3/100\n9/9 [==============================] - 7s 784ms/step - loss: 0.9784 - val_loss: 0.9454\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 4/100\n9/9 [==============================] - 6s 711ms/step - loss: 0.9238 - val_loss: 0.8984\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 5/100\n9/9 [==============================] - 6s 714ms/step - loss: 0.8798 - val_loss: 0.8603\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 6/100\n9/9 [==============================] - 6s 715ms/step - loss: 0.8441 - val_loss: 0.8286\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 7/100\n9/9 [==============================] - 7s 723ms/step - loss: 0.8147 - val_loss: 0.8023\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 8/100\n9/9 [==============================] - 7s 767ms/step - loss: 0.7899 - val_loss: 0.7799\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 9/100\n9/9 [==============================] - 7s 735ms/step - loss: 0.7686 - val_loss: 0.7602\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 10/100\n9/9 [==============================] - 6s 719ms/step - loss: 0.7490 - val_loss: 0.7392\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 11/100\n9/9 [==============================] - 6s 722ms/step - loss: 0.7259 - val_loss: 0.7111\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 12/100\n9/9 [==============================] - 7s 730ms/step - loss: 0.6946 - val_loss: 0.6712\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 13/100\n9/9 [==============================] - 6s 717ms/step - loss: 0.6581 - val_loss: 0.6386\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 14/100\n9/9 [==============================] - 7s 764ms/step - loss: 0.6278 - val_loss: 0.6103\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 15/100\n9/9 [==============================] - 7s 745ms/step - loss: 0.6020 - val_loss: 0.5864\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 16/100\n9/9 [==============================] - 6s 715ms/step - loss: 0.5808 - val_loss: 0.5673\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 17/100\n9/9 [==============================] - 7s 824ms/step - loss: 0.5635 - val_loss: 0.5518\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 18/100\n9/9 [==============================] - 7s 817ms/step - loss: 0.5492 - val_loss: 0.5389\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 19/100\n9/9 [==============================] - 8s 875ms/step - loss: 0.5373 - val_loss: 0.5281\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 20/100\n9/9 [==============================] - 8s 849ms/step - loss: 0.5273 - val_loss: 0.5188\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 21/100\n9/9 [==============================] - 8s 845ms/step - loss: 0.5183 - val_loss: 0.5102\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 22/100\n9/9 [==============================] - 7s 803ms/step - loss: 0.5104 - val_loss: 0.5027\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 23/100\n9/9 [==============================] - 7s 736ms/step - loss: 0.5032 - val_loss: 0.4959\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 24/100\n9/9 [==============================] - 7s 734ms/step - loss: 0.4968 - val_loss: 0.4900\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 25/100\n9/9 [==============================] - 7s 746ms/step - loss: 0.4908 - val_loss: 0.4844\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 26/100\n9/9 [==============================] - 7s 733ms/step - loss: 0.4855 - val_loss: 0.4794\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 27/100\n9/9 [==============================] - 7s 749ms/step - loss: 0.4808 - val_loss: 0.4751\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 28/100\n9/9 [==============================] - 7s 781ms/step - loss: 0.4766 - val_loss: 0.4710\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 29/100\n9/9 [==============================] - 6s 713ms/step - loss: 0.4727 - val_loss: 0.4675\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 30/100\n9/9 [==============================] - 6s 719ms/step - loss: 0.4692 - val_loss: 0.4641\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 31/100\n9/9 [==============================] - 6s 713ms/step - loss: 0.4659 - val_loss: 0.4608\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 32/100\n9/9 [==============================] - 6s 717ms/step - loss: 0.4628 - val_loss: 0.4580\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 33/100\n9/9 [==============================] - 7s 734ms/step - loss: 0.4599 - val_loss: 0.4551\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 34/100\n9/9 [==============================] - 12s 1s/step - loss: 0.4569 - val_loss: 0.4522\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 35/100\n9/9 [==============================] - 9s 1s/step - loss: 0.4541 - val_loss: 0.4495\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 36/100\n9/9 [==============================] - 7s 833ms/step - loss: 0.4512 - val_loss: 0.4466\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 37/100\n9/9 [==============================] - 8s 909ms/step - loss: 0.4480 - val_loss: 0.4433\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 38/100\n9/9 [==============================] - 8s 942ms/step - loss: 0.4445 - val_loss: 0.4398\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 39/100\n9/9 [==============================] - 8s 862ms/step - loss: 0.4408 - val_loss: 0.4360\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 40/100\n9/9 [==============================] - 7s 786ms/step - loss: 0.4364 - val_loss: 0.4324\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 41/100\n9/9 [==============================] - 7s 788ms/step - loss: 0.4322 - val_loss: 0.4285\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 42/100\n9/9 [==============================] - 7s 723ms/step - loss: 0.4279 - val_loss: 0.4245\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 43/100\n9/9 [==============================] - 8s 876ms/step - loss: 0.4237 - val_loss: 0.4202\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 44/100\n9/9 [==============================] - 8s 880ms/step - loss: 0.4195 - val_loss: 0.4163\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 45/100\n9/9 [==============================] - 8s 899ms/step - loss: 0.4152 - val_loss: 0.4123\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 46/100\n9/9 [==============================] - 8s 857ms/step - loss: 0.4108 - val_loss: 0.4076\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 47/100\n9/9 [==============================] - 8s 835ms/step - loss: 0.4059 - val_loss: 0.4031\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 48/100\n9/9 [==============================] - 7s 775ms/step - loss: 0.4013 - val_loss: 0.3986\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 49/100\n9/9 [==============================] - 7s 817ms/step - loss: 0.3969 - val_loss: 0.3946\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 50/100\n9/9 [==============================] - 7s 792ms/step - loss: 0.3928 - val_loss: 0.3909\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 51/100\n9/9 [==============================] - 7s 803ms/step - loss: 0.3889 - val_loss: 0.3873\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 52/100\n9/9 [==============================] - 7s 788ms/step - loss: 0.3855 - val_loss: 0.3841\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 53/100\n9/9 [==============================] - 9s 1s/step - loss: 0.3823 - val_loss: 0.3814\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 54/100\n9/9 [==============================] - 7s 785ms/step - loss: 0.3794 - val_loss: 0.3785\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 55/100\n9/9 [==============================] - 7s 729ms/step - loss: 0.3766 - val_loss: 0.3762\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 56/100\n9/9 [==============================] - 7s 739ms/step - loss: 0.3739 - val_loss: 0.3734\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 57/100\n9/9 [==============================] - 7s 738ms/step - loss: 0.3714 - val_loss: 0.3713\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 58/100\n9/9 [==============================] - 7s 807ms/step - loss: 0.3689 - val_loss: 0.3689\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 59/100\n9/9 [==============================] - 7s 734ms/step - loss: 0.3666 - val_loss: 0.3668\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 60/100\n9/9 [==============================] - 7s 731ms/step - loss: 0.3644 - val_loss: 0.3648\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 61/100\n9/9 [==============================] - 7s 730ms/step - loss: 0.3622 - val_loss: 0.3624\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 62/100\n9/9 [==============================] - 7s 822ms/step - loss: 0.3599 - val_loss: 0.3605\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 63/100\n9/9 [==============================] - 7s 754ms/step - loss: 0.3578 - val_loss: 0.3581\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 64/100\n9/9 [==============================] - 7s 804ms/step - loss: 0.3554 - val_loss: 0.3561\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 65/100\n9/9 [==============================] - 7s 758ms/step - loss: 0.3534 - val_loss: 0.3542\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 66/100\n9/9 [==============================] - 8s 873ms/step - loss: 0.3511 - val_loss: 0.3519\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 67/100\n9/9 [==============================] - 7s 816ms/step - loss: 0.3490 - val_loss: 0.3501\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 68/100\n9/9 [==============================] - 7s 737ms/step - loss: 0.3470 - val_loss: 0.3480\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 69/100\n9/9 [==============================] - 7s 723ms/step - loss: 0.3448 - val_loss: 0.3458\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 70/100\n9/9 [==============================] - 6s 714ms/step - loss: 0.3426 - val_loss: 0.3440\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 71/100\n9/9 [==============================] - 6s 713ms/step - loss: 0.3408 - val_loss: 0.3423\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 72/100\n9/9 [==============================] - 6s 714ms/step - loss: 0.3389 - val_loss: 0.3404\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 73/100\n9/9 [==============================] - 7s 742ms/step - loss: 0.3372 - val_loss: 0.3389\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 74/100\n9/9 [==============================] - 7s 800ms/step - loss: 0.3354 - val_loss: 0.3369\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 75/100\n9/9 [==============================] - 8s 937ms/step - loss: 0.3337 - val_loss: 0.3356\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 76/100\n9/9 [==============================] - 7s 743ms/step - loss: 0.3321 - val_loss: 0.3337\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 77/100\n9/9 [==============================] - 7s 723ms/step - loss: 0.3305 - val_loss: 0.3322\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 78/100\n9/9 [==============================] - 6s 717ms/step - loss: 0.3289 - val_loss: 0.3307\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 79/100\n9/9 [==============================] - 6s 717ms/step - loss: 0.3275 - val_loss: 0.3294\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 80/100\n9/9 [==============================] - 8s 861ms/step - loss: 0.3259 - val_loss: 0.3279\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 81/100\n9/9 [==============================] - 7s 756ms/step - loss: 0.3244 - val_loss: 0.3265\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 82/100\n9/9 [==============================] - 7s 737ms/step - loss: 0.3229 - val_loss: 0.3250\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 83/100\n9/9 [==============================] - 6s 719ms/step - loss: 0.3213 - val_loss: 0.3236\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 84/100\n9/9 [==============================] - 6s 717ms/step - loss: 0.3199 - val_loss: 0.3221\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 85/100\n9/9 [==============================] - 6s 720ms/step - loss: 0.3184 - val_loss: 0.3207\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 86/100\n9/9 [==============================] - 6s 719ms/step - loss: 0.3170 - val_loss: 0.3192\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 87/100\n9/9 [==============================] - 7s 728ms/step - loss: 0.3153 - val_loss: 0.3178\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 88/100\n9/9 [==============================] - 7s 736ms/step - loss: 0.3139 - val_loss: 0.3165\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 89/100\n9/9 [==============================] - 6s 715ms/step - loss: 0.3126 - val_loss: 0.3152\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 90/100\n9/9 [==============================] - 6s 720ms/step - loss: 0.3114 - val_loss: 0.3140\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 91/100\n9/9 [==============================] - 7s 731ms/step - loss: 0.3100 - val_loss: 0.3127\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 92/100\n9/9 [==============================] - 7s 734ms/step - loss: 0.3085 - val_loss: 0.3113\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 93/100\n9/9 [==============================] - 7s 726ms/step - loss: 0.3072 - val_loss: 0.3101\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 94/100\n9/9 [==============================] - 7s 727ms/step - loss: 0.3062 - val_loss: 0.3091\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 95/100\n9/9 [==============================] - 7s 726ms/step - loss: 0.3049 - val_loss: 0.3079\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 96/100\n9/9 [==============================] - 7s 733ms/step - loss: 0.3037 - val_loss: 0.3065\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 97/100\n9/9 [==============================] - 6s 722ms/step - loss: 0.3024 - val_loss: 0.3053\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 98/100\n9/9 [==============================] - 7s 722ms/step - loss: 0.3012 - val_loss: 0.3042\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 99/100\n9/9 [==============================] - 7s 730ms/step - loss: 0.3000 - val_loss: 0.3031\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 100/100\n9/9 [==============================] - 7s 722ms/step - loss: 0.2989 - val_loss: 0.3018\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nCompleted 1 reconstruction and encoding, saved test data to assess performance\nStarting on index: 16\nEpoch 1/100\n9/9 [==============================] - 6s 714ms/step - loss: 1.1664 - val_loss: 1.1320\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 2/100\n9/9 [==============================] - 6s 690ms/step - loss: 1.1051 - val_loss: 1.0757\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 3/100\n9/9 [==============================] - 6s 699ms/step - loss: 1.0580 - val_loss: 1.0400\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 4/100\n9/9 [==============================] - 6s 697ms/step - loss: 1.0291 - val_loss: 1.0166\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 5/100\n9/9 [==============================] - 6s 694ms/step - loss: 1.0076 - val_loss: 0.9953\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 6/100\n9/9 [==============================] - 6s 710ms/step - loss: 0.9854 - val_loss: 0.9715\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 7/100\n9/9 [==============================] - 6s 694ms/step - loss: 0.9609 - val_loss: 0.9465\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 8/100\n9/9 [==============================] - 6s 704ms/step - loss: 0.9364 - val_loss: 0.9231\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 9/100\n9/9 [==============================] - 6s 696ms/step - loss: 0.9134 - val_loss: 0.9006\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 10/100\n9/9 [==============================] - 6s 702ms/step - loss: 0.8909 - val_loss: 0.8783\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 11/100\n9/9 [==============================] - 6s 705ms/step - loss: 0.8682 - val_loss: 0.8555\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nEpoch 12/100\n8/9 [=========================>....] - ETA: 0s - loss: 0.8463\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b"
],
[
"def mean_squared_error(reduced_dimensions, model_name, patient_num, save_errors=False):\n \"\"\"\n Computes the mean squared error of the reconstructed signal against the original signal for each lead for each of the patient_num\n Each signal's dimensions are reduced from 100 to 'reduced_dimensions', then reconstructed to obtain the reconstructed signal\n\n :param reduced_dimensions: number of dimensions the file was originally reduced to\n :param model_name: \"lstm, vae, ae, pca, test\"\n :return: dictionary of patient_index -> length n array of MSE for each heartbeat (i.e. MSE of 100x4 arrays)\n \"\"\"\n print(\"calculating mse for file index {} on the reconstructed {} model\".format(patient_num, model_name))\n original_signals = np.load(\n os.path.join(\"Working_Data\", \"Training_Subset\", \"Normalized\", \"ten_hbs\", \"Normalized_Fixed_Dim_HBs_Idx{}.npy\".format(str(patient_num))))\n\n print(\"original normalized signal\")\n # print(original_signals[0, :,:])\n # print(np.mean(original_signals[0,:,:]))\n # print(np.var(original_signals[0, :, :]))\n # print(np.linalg.norm(original_signals[0,:,:]))\n # print([np.linalg.norm(i) for i in original_signals[0,:,:].flatten()])\n\n\n reconstructed_signals = np.load(os.path.join(\"Working_Data\",\"Training_Subset\", \"Model_Output\",\n \"reconstructed_10hb_cae_{}.npy\").format(str(patient_num)))\n # compute mean squared error for each heartbeat\n # mse = (np.square(original_signals - reconstructed_signals) / (np.linalg.norm(original_signals))).mean(axis=1).mean(axis=1)\n # mse = (np.square(original_signals - reconstructed_signals) / (np.square(original_signals) + np.square(reconstructed_signals))).mean(axis=1).mean(axis=1)\n\n mse = np.zeros(np.shape(original_signals)[0])\n for i in range(np.shape(original_signals)[0]):\n mse[i] = (np.linalg.norm(original_signals[i,:,:] - reconstructed_signals[i,:,:]) ** 2) / (np.linalg.norm(original_signals[i,:,:]) ** 2)\n # orig_flat = original_signals[i,:,:].flatten()\n # recon_flat = reconstructed_signals[i,:,:].flatten()\n # mse[i] = sklearn_mse(orig_flat, recon_flat)\n # my_mse = mse[i]\n\n # plt.plot([i for i in range(np.shape(mse)[0])], mse)\n # plt.show()\n\n if save_errors:\n np.save(\n os.path.join(\"Working_Data\", \"{}_errors_{}d_Idx{}.npy\".format(model_name, reduced_dimensions, patient_num)), mse)\n # print(list(mse))\n\n # return np.array([err for err in mse if 1 == 1 and err < 5 and 0 == 0 and 3 < 4])\n return mse",
"_____no_output_____"
],
[
"def windowed_mse_over_time(patient_num, model_name, dimension_num):\n errors = mean_squared_error(dimension_num, model_name, patient_num, False)\n\n # window the errors - assume 500 samples ~ 5 min\n window_duration = 250\n windowed_errors = []\n for i in range(0, len(errors) - window_duration, window_duration):\n windowed_errors.append(np.mean(errors[i:i+window_duration]))\n\n sample_idcs = [i for i in range(len(windowed_errors))]\n print(windowed_errors)\n plt.plot(sample_idcs, windowed_errors)\n plt.title(\"5-min Windowed MSE\" + str(patient_num))\n plt.xlabel(\"Window Index\")\n plt.ylabel(\"Relative MSE\")\n plt.show()\n\n\n # np.save(f\"Working_Data/windowed_mse_{dimension_num}d_Idx{patient_num}.npy\", windowed_errors)",
"_____no_output_____"
],
[
"windowed_mse_over_time(1,\"abc\",10)",
"calculating mse for file index 1 on the reconstructed abc model\noriginal normalized signal\n[0.14156092541205495, 0.12069838606490167, 0.11476877337544941, 0.10755301753206598, 0.0643757580002823, 0.18476382650404158, 0.3032713164345003, 0.31138377123331595, 0.3625266461386575, 0.36354417970353575, 0.316255777127343, 0.33135989810073524, 0.39180671485961605, 0.39716030146613107, 0.3803097963441608, 0.4030001293594089, 0.3820503543084321, 0.3865000524709579, 0.38972508277616214]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0009d5398ddafa5402c0865af43fc2773a267cc | 190,653 | ipynb | Jupyter Notebook | exercise_2.ipynb | deepak223098/Computer_Vision_Example | d477c1ef04f5e6eb58f078da03efce7a2c63f88b | [
"Apache-2.0"
] | 1 | 2020-08-10T05:32:41.000Z | 2020-08-10T05:32:41.000Z | exercise_2.ipynb | deepak223098/Computer_Vision_Example | d477c1ef04f5e6eb58f078da03efce7a2c63f88b | [
"Apache-2.0"
] | null | null | null | exercise_2.ipynb | deepak223098/Computer_Vision_Example | d477c1ef04f5e6eb58f078da03efce7a2c63f88b | [
"Apache-2.0"
] | null | null | null | 332.148084 | 40,428 | 0.932338 | [
[
[
"# basic operation on image",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimpath = r\"D:/Study/example_ml/computer_vision_example/cv_exercise/opencv-master/samples/data/messi5.jpg\"\nimg = cv2.imread(impath)\nprint(img.shape)\nprint(img.size)\nprint(img.dtype)\nb,g,r = cv2.split(img)\nimg = cv2.merge((b,g,r))\ncv2.imshow(\"image\",img)\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"# copy and paste",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimpath = r\"D:/Study/example_ml/computer_vision_example/cv_exercise/opencv-master/samples/data/messi5.jpg\"\nimg = cv2.imread(impath)\n'''b,g,r = cv2.split(img)\nimg = cv2.merge((b,g,r))'''\nball = img[280:340,330:390]\nimg[273:333,100:160] = ball\ncv2.imshow(\"image\",img)\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"# merge two imge",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimpath = r\"D:/Study/example_ml/computer_vision_example/cv_exercise/opencv-master/samples/data/messi5.jpg\"\nimpath1 = r\"D:/Study/example_ml/computer_vision_example/cv_exercise/opencv-master/samples/data/opencv-logo.png\"\nimg = cv2.imread(impath)\nimg1 = cv2.imread(impath1)\nimg = cv2.resize(img, (512,512))\nimg1 = cv2.resize(img1, (512,512))\n#new_img = cv2.add(img,img1)\nnew_img = cv2.addWeighted(img,0.1,img1,0.8,1)\ncv2.imshow(\"new_image\",new_img)\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"# bitwise operation ",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimg1 = np.zeros([250,500,3],np.uint8)\nimg1 = cv2.rectangle(img1,(200,0),(300,100),(255,255,255),-1)\nimg2 = np.full((250, 500, 3), 255, dtype=np.uint8)\nimg2 = cv2.rectangle(img2, (0, 0), (250, 250), (0, 0, 0), -1)\n#bit_and = cv2.bitwise_and(img2,img1)\n#bit_or = cv2.bitwise_or(img2,img1)\n#bit_xor = cv2.bitwise_xor(img2,img1)\nbit_not = cv2.bitwise_not(img2)\n#cv2.imshow(\"bit_and\",bit_and)\n#cv2.imshow(\"bit_or\",bit_or)\n#cv2.imshow(\"bit_xor\",bit_xor)\ncv2.imshow(\"bit_not\",bit_not)\ncv2.imshow(\"img1\",img1) \ncv2.imshow(\"img2\",img2)\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"# simple thresholding ",
"_____no_output_____"
],
[
"#### THRESH_BINARY",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimg = cv2.imread('gradient.jpg',0)\n_,th1 = cv2.threshold(img,127,255,cv2.THRESH_BINARY) #check every pixel with 127\ncv2.imshow(\"img\",img)\ncv2.imshow(\"th1\",th1)\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"#### THRESH_BINARY_INV",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimg = cv2.imread('gradient.jpg',0)\n_,th1 = cv2.threshold(img,127,255,cv2.THRESH_BINARY)\n_,th2 = cv2.threshold(img,127,255,cv2.THRESH_BINARY_INV) #check every pixel with 127\ncv2.imshow(\"img\",img)\ncv2.imshow(\"th1\",th1)\ncv2.imshow(\"th2\",th2)\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"#### THRESH_TRUNC",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimg = cv2.imread('gradient.jpg',0)\n_,th1 = cv2.threshold(img,127,255,cv2.THRESH_BINARY)\n_,th2 = cv2.threshold(img,255,255,cv2.THRESH_TRUNC) #check every pixel with 127\ncv2.imshow(\"img\",img)\ncv2.imshow(\"th1\",th1)\ncv2.imshow(\"th2\",th2)\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"#### THRESH_TOZERO",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimg = cv2.imread('gradient.jpg',0)\n\n_,th1 = cv2.threshold(img,127,255,cv2.THRESH_BINARY)\n_,th2 = cv2.threshold(img,127,255,cv2.THRESH_TOZERO) #check every pixel with 127\n_,th3 = cv2.threshold(img,127,255,cv2.THRESH_TOZERO_INV) #check every pixel with 127\n\ncv2.imshow(\"img\",img)\ncv2.imshow(\"th1\",th1)\ncv2.imshow(\"th2\",th2)\ncv2.imshow(\"th3\",th3)\n\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"# Adaptive Thresholding\n##### it will calculate the threshold for smaller region of iamge .so we get different thresholding value for different region of same image",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\nimg = cv2.imread('sudoku1.jpg')\nimg = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)\n_,th1 = cv2.threshold(img,127,255,cv2.THRESH_BINARY)\nth2 = cv2.adaptiveThreshold(img,255,cv2.ADAPTIVE_THRESH_MEAN_C,\n cv2.THRESH_BINARY,11,2)\nth3 = cv2.adaptiveThreshold(img,255,cv2.ADAPTIVE_THRESH_GAUSSIAN_C,\n cv2.THRESH_BINARY,11,2)\n\n\ncv2.imshow(\"img\",img)\ncv2.imshow(\"THRESH_BINARY\",th1)\ncv2.imshow(\"ADAPTIVE_THRESH_MEAN_C\",th2)\ncv2.imshow(\"ADAPTIVE_THRESH_GAUSSIAN_C\",th3)\n\ncv2.waitKey(0)\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"# Morphological Transformations\n\n#### Morphological Transformations are some simple operation based on the image shape. Morphological Transformations are normally performed on binary images.\n#### A kernal tells you how to change the value of any given pixel by combining it with different amounts of the neighbouring pixels.",
"_____no_output_____"
]
],
[
[
"import cv2\n%matplotlib notebook\n%matplotlib inline\nfrom matplotlib import pyplot as plt\nimg = cv2.imread(\"hsv_ball.jpg\",cv2.IMREAD_GRAYSCALE)\n_,mask = cv2.threshold(img, 220,255,cv2.THRESH_BINARY_INV)\ntitles = ['images',\"mask\"]\nimages = [img,mask]\nfor i in range(2):\n plt.subplot(1,2,i+1)\n plt.imshow(images[i],\"gray\")\n plt.title(titles[i])\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Morphological Transformations using erosion",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\n%matplotlib notebook\n%matplotlib inline\nfrom matplotlib import pyplot as plt\nimg = cv2.imread(\"hsv_ball.jpg\",cv2.IMREAD_GRAYSCALE)\n_,mask = cv2.threshold(img, 220,255,cv2.THRESH_BINARY_INV)\nkernal = np.ones((2,2),np.uint8)\ndilation = cv2.dilate(mask,kernal,iterations = 3)\nerosion = cv2.erode(mask,kernal,iterations=1)\ntitles = ['images',\"mask\",\"dilation\",\"erosion\"]\nimages = [img,mask,dilation,erosion]\nfor i in range(len(titles)):\n plt.subplot(2,2,i+1)\n plt.imshow(images[i],\"gray\")\n plt.title(titles[i])\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Morphological Transformations using opening morphological operation\n\n##### morphologyEx . Will use erosion operation first then dilation on the image",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\n%matplotlib notebook\n%matplotlib inline\nfrom matplotlib import pyplot as plt\nimg = cv2.imread(\"hsv_ball.jpg\",cv2.IMREAD_GRAYSCALE)\n_,mask = cv2.threshold(img, 220,255,cv2.THRESH_BINARY_INV)\nkernal = np.ones((5,5),np.uint8)\ndilation = cv2.dilate(mask,kernal,iterations = 3)\nerosion = cv2.erode(mask,kernal,iterations=1)\nopening = cv2.morphologyEx(mask,cv2.MORPH_OPEN,kernal)\ntitles = ['images',\"mask\",\"dilation\",\"erosion\",\"opening\"]\nimages = [img,mask,dilation,erosion,opening]\nfor i in range(len(titles)):\n plt.subplot(2,3,i+1)\n plt.imshow(images[i],\"gray\")\n plt.title(titles[i])\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Morphological Transformations using closing morphological operation\n\n##### morphologyEx . Will use dilation operation first then erosion on the image",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\n%matplotlib notebook\n%matplotlib inline\nfrom matplotlib import pyplot as plt\nimg = cv2.imread(\"hsv_ball.jpg\",cv2.IMREAD_GRAYSCALE)\n_,mask = cv2.threshold(img, 220,255,cv2.THRESH_BINARY_INV)\nkernal = np.ones((5,5),np.uint8)\ndilation = cv2.dilate(mask,kernal,iterations = 3)\nerosion = cv2.erode(mask,kernal,iterations=1)\nopening = cv2.morphologyEx(mask,cv2.MORPH_OPEN,kernal)\nclosing = cv2.morphologyEx(mask,cv2.MORPH_CLOSE,kernal)\ntitles = ['images',\"mask\",\"dilation\",\"erosion\",\"opening\",\"closing\"]\nimages = [img,mask,dilation,erosion,opening,closing]\nfor i in range(len(titles)):\n plt.subplot(2,3,i+1)\n plt.imshow(images[i],\"gray\")\n plt.title(titles[i])\n plt.xticks([])\n plt.yticks([])\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Morphological Transformations other than opening and closing morphological operation\n#### MORPH_GRADIENT will give the difference between dilation and erosion\n#### top_hat will give the difference between input image and opening image",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\n%matplotlib notebook\n%matplotlib inline\nfrom matplotlib import pyplot as plt\nimg = cv2.imread(\"hsv_ball.jpg\",cv2.IMREAD_GRAYSCALE)\n_,mask = cv2.threshold(img, 220,255,cv2.THRESH_BINARY_INV)\nkernal = np.ones((5,5),np.uint8)\ndilation = cv2.dilate(mask,kernal,iterations = 3)\nerosion = cv2.erode(mask,kernal,iterations=1)\nopening = cv2.morphologyEx(mask,cv2.MORPH_OPEN,kernal)\nclosing = cv2.morphologyEx(mask,cv2.MORPH_CLOSE,kernal)\nmorphlogical_gradient = cv2.morphologyEx(mask,cv2.MORPH_GRADIENT,kernal)\ntop_hat = cv2.morphologyEx(mask,cv2.MORPH_TOPHAT,kernal)\ntitles = ['images',\"mask\",\"dilation\",\"erosion\",\"opening\",\n \"closing\",\"morphlogical_gradient\",\"top_hat\"]\nimages = [img,mask,dilation,erosion,opening,\n closing,morphlogical_gradient,top_hat]\nfor i in range(len(titles)):\n plt.subplot(2,4,i+1)\n plt.imshow(images[i],\"gray\")\n plt.title(titles[i])\n plt.xticks([])\n plt.yticks([])\nplt.show()",
"_____no_output_____"
],
[
"import cv2\nimport numpy as np\n%matplotlib notebook\n%matplotlib inline\nfrom matplotlib import pyplot as plt\nimg = cv2.imread(\"HappyFish.jpg\",cv2.IMREAD_GRAYSCALE)\n_,mask = cv2.threshold(img, 220,255,cv2.THRESH_BINARY_INV)\nkernal = np.ones((5,5),np.uint8)\ndilation = cv2.dilate(mask,kernal,iterations = 3)\nerosion = cv2.erode(mask,kernal,iterations=1)\nopening = cv2.morphologyEx(mask,cv2.MORPH_OPEN,kernal)\nclosing = cv2.morphologyEx(mask,cv2.MORPH_CLOSE,kernal)\nMORPH_GRADIENT = cv2.morphologyEx(mask,cv2.MORPH_GRADIENT,kernal)\ntop_hat = cv2.morphologyEx(mask,cv2.MORPH_TOPHAT,kernal)\ntitles = ['images',\"mask\",\"dilation\",\"erosion\",\"opening\",\n \"closing\",\"MORPH_GRADIENT\",\"top_hat\"]\nimages = [img,mask,dilation,erosion,opening,\n closing,MORPH_GRADIENT,top_hat]\nfor i in range(len(titles)):\n plt.subplot(2,4,i+1)\n plt.imshow(images[i],\"gray\")\n plt.title(titles[i])\n plt.xticks([])\n plt.yticks([])\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d000a660b0e9fa13fa75686a8147c7afbccbc039 | 34,254 | ipynb | Jupyter Notebook | Untitled1.ipynb | archit120/lingatagger | cb3d0e262900dba1fd1ead0a37fad531e37cff9f | [
"Apache-2.0"
] | 1 | 2019-06-29T10:59:22.000Z | 2019-06-29T10:59:22.000Z | Untitled1.ipynb | archit120/lingatagger | cb3d0e262900dba1fd1ead0a37fad531e37cff9f | [
"Apache-2.0"
] | null | null | null | Untitled1.ipynb | archit120/lingatagger | cb3d0e262900dba1fd1ead0a37fad531e37cff9f | [
"Apache-2.0"
] | null | null | null | 38.925 | 152 | 0.476791 | [
[
[
"Create a list of valid Hindi literals",
"_____no_output_____"
]
],
[
[
"a = list(set(list(\"ऀँंःऄअआइईउऊऋऌऍऎएऐऑऒओऔकखगघङचछजझञटठडढणतथदधनऩपफबभमयरऱलळऴवशषसहऺऻ़ऽािीुूृॄॅॆेैॉॊोौ्ॎॏॐ॒॑॓॔ॕॖॗक़ख़ग़ज़ड़ढ़फ़य़ॠॡॢॣ।॥॰ॱॲॳॴॵॶॷॸॹॺॻॼॽॾॿ-\")))\n",
"_____no_output_____"
],
[
"len(genderListCleared),len(set(genderListCleared))",
"_____no_output_____"
],
[
"genderListCleared = list(set(genderListCleared))",
"_____no_output_____"
],
[
"mCount = 0\nfCount = 0\nnCount = 0\nfor item in genderListCleared:\n if item[1] == 'm':\n mCount+=1\n elif item[1] == 'f':\n fCount+=1\n elif item[1] == 'none':\n nCount+=1",
"_____no_output_____"
],
[
"mCount,fCount,nCount,len(genderListCleared)-mCount-fCount-nCount",
"_____no_output_____"
],
[
"with open('genderListCleared', 'wb') as fp:\n pickle.dump(genderListCleared, fp)\n",
"_____no_output_____"
],
[
"with open('genderListCleared', 'rb') as fp:\n genderListCleared = pickle.load(fp)\n",
"_____no_output_____"
],
[
"genderListNoNone= []\nfor item in genderListCleared:\n if item[1] == 'm':\n genderListNoNone.append(item)\n elif item[1] == 'f':\n genderListNoNone.append(item)\n elif item[1] == 'any':\n genderListNoNone.append(item)\n",
"_____no_output_____"
],
[
"with open('genderListNoNone', 'wb') as fp:\n pickle.dump(genderListNoNone, fp)\n",
"_____no_output_____"
],
[
"with open('genderListNoNone', 'rb') as fp:\n genderListNoNone = pickle.load(fp)\n",
"_____no_output_____"
],
[
"noneWords = list(set(genderListCleared)-set(genderListNoNone))",
"_____no_output_____"
],
[
"noneWords = set([x[0] for x in noneWords])",
"_____no_output_____"
],
[
"import lingatagger.genderlist as gndrlist\nimport lingatagger.tokenizer as tok\nfrom lingatagger.tagger import *\n\ngenders2 = gndrlist.drawlist()\ngenderList2 = []\nfor i in genders2:\n x = i.split(\"\\t\")\n if type(numericTagger(x[0])[0]) != tuple:\n count = 0\n for ch in list(x[0]):\n if ch not in a:\n count+=1\n if count == 0:\n if len(x)>=3:\n genderList2.append((x[0],'any'))\n else:\n genderList2.append((x[0],x[1]))\n",
"_____no_output_____"
],
[
"genderList2.sort()\ngenderList2Cleared = genderList2\nfor ind in range(0, len(genderList2Cleared)-1):\n if genderList2Cleared[ind][0] == genderList2Cleared[ind+1][0]:\n genderList2Cleared[ind] = genderList2Cleared[ind][0], 'any'\n genderList2Cleared[ind+1] = genderList2Cleared[ind][0], 'any'",
"_____no_output_____"
],
[
"genderList2Cleared = list(set(genderList2Cleared))",
"_____no_output_____"
],
[
"mCount2 = 0\nfCount2 = 0\nfor item in genderList2Cleared:\n if item[1] == 'm':\n mCount2+=1\n elif item[1] == 'f':\n fCount2+=1",
"_____no_output_____"
],
[
"mCount2,fCount2,len(genderList2Cleared)-mCount2-fCount2",
"_____no_output_____"
],
[
"with open('genderList2Cleared', 'wb') as fp:\n pickle.dump(genderList2Cleared, fp)\n",
"_____no_output_____"
],
[
"with open('genderList2Cleared', 'rb') as fp:\n genderList2Cleared = pickle.load(fp)\n",
"_____no_output_____"
],
[
"genderList2Matched = []\nfor item in genderList2Cleared:\n if item[0] in noneWords:\n continue\n genderList2Matched.append(item)",
"_____no_output_____"
],
[
"len(genderList2Cleared)-len(genderList2Matched)",
"_____no_output_____"
],
[
"with open('genderList2Matched', 'wb') as fp:\n pickle.dump(genderList2Matched, fp)\n",
"_____no_output_____"
],
[
"mergedList = []\nfor item in genderList2Cleared:\n mergedList.append((item[0], item[1]))\nfor item in genderListNoNone:\n mergedList.append((item[0], item[1]))\nmergedList.sort()",
"_____no_output_____"
],
[
"for ind in range(0, len(mergedList)-1):\n if mergedList[ind][0] == mergedList[ind+1][0]:\n fgend = 'any'\n if mergedList[ind][1] == 'm' or mergedList[ind+1][1] == 'm':\n fgend = 'm'\n elif mergedList[ind][1] == 'f' or mergedList[ind+1][1] == 'f':\n if fgend == 'm':\n fgend = 'any'\n else:\n fgend = 'f'\n else:\n fgend = 'any'\n mergedList[ind] = mergedList[ind][0], fgend\n mergedList[ind+1] = mergedList[ind][0], fgend\n\nmergedList = list(set(mergedList))",
"_____no_output_____"
],
[
"mCount3 = 0\nfCount3 = 0\nfor item in mergedList:\n if item[1] == 'm':\n mCount3+=1\n elif item[1] == 'f':\n fCount3+=1",
"_____no_output_____"
],
[
"mCount3,fCount3,len(mergedList)-mCount3-fCount3",
"_____no_output_____"
],
[
"with open('mergedList', 'wb') as fp:\n pickle.dump(mergedList, fp)\n",
"_____no_output_____"
],
[
"with open('mergedList', 'rb') as fp:\n mergedList = pickle.load(fp)\n",
"_____no_output_____"
],
[
"np.zeros(18, dtype=\"int\")\n",
"_____no_output_____"
],
[
"from keras.models import Sequential\nfrom keras.layers import Dense, Dropout\nfrom keras.layers import Embedding\nfrom keras.layers import Conv1D, GlobalAveragePooling1D, MaxPooling1D\nfrom keras.layers import Dense, Conv2D, Flatten\nfrom sklearn.feature_extraction.text import CountVectorizer\nimport numpy as np\nimport lingatagger.genderlist as gndrlist\nimport lingatagger.tokenizer as tok\nfrom lingatagger.tagger import *\nimport re\nimport heapq\n\ndef encodex(text):\n s = list(text)\n \n indices = []\n for i in s:\n indices.append(a.index(i))\n encoded = np.zeros(18, dtype=\"int\")\n #print(len(a)+1)\n k = 0\n for i in indices:\n encoded[k] = i\n k = k + 1\n for i in range(18-len(list(s))):\n encoded[k+i] = len(a)\n return encoded\n\ndef encodey(text):\n if text == \"f\":\n return [1,0,0]\n elif text == \"m\":\n return [0,0,1]\n\n else:\n return [0,1,0] \n\ndef genderdecode(genderTag):\n \"\"\"\n one-hot decoding for the gender tag predicted by the classfier\n Dimension = 2.\n \"\"\"\n genderTag = list(genderTag[0])\n index = genderTag.index(heapq.nlargest(1, genderTag)[0])\n if index == 0:\n return 'f'\n if index == 2:\n return 'm'\n if index == 1:\n return 'any'\n",
"_____no_output_____"
],
[
"\nx_train = []\ny_train = []\nfor i in genderListNoNone:\n if len(i[0]) > 18:\n continue\n x_train.append(encodex(i[0]))\n y_train.append(encodey(i[1]))\n \n\nx_test = []\ny_test = []\nfor i in genderList2Matched:\n if len(i[0]) > 18:\n continue\n x_test.append(encodex(i[0]))\n y_test.append(encodey(i[1]))\n \n \n\nx_merged = []\ny_merged = []\nfor i in mergedList:\n if len(i[0]) > 18:\n continue\n x_merged.append(encodex(i[0]))\n y_merged.append(encodey(i[1]))",
"_____no_output_____"
],
[
"\nX_train = np.array(x_train)\nY_train = np.array(y_train)\n\nX_test = np.array(x_test)\nY_test = np.array(y_test)\n\nX_merged = np.array(x_merged)\nY_merged = np.array(y_merged)\n\nwith open('X_train', 'wb') as fp:\n pickle.dump(X_train, fp)\n\nwith open('Y_train', 'wb') as fp:\n pickle.dump(Y_train, fp)\n\nwith open('X_test', 'wb') as fp:\n pickle.dump(X_test, fp)\n\nwith open('Y_test', 'wb') as fp:\n pickle.dump(Y_test, fp)\n",
"_____no_output_____"
],
[
"from keras.models import Sequential\nfrom keras.layers import Dense, Dropout\nfrom keras.layers import Embedding\nfrom keras.layers import LSTM\n\nmax_features = len(a)+1\n\nfor loss_f in ['categorical_crossentropy']:\n for opt in ['rmsprop','adam','nadam','sgd']:\n for lstm_len in [32,64,128,256]:\n for dropout in [0.4,0.45,0.5,0.55,0.6]:\n model = Sequential()\n model.add(Embedding(max_features, output_dim=18))\n model.add(LSTM(lstm_len))\n model.add(Dropout(dropout))\n model.add(Dense(3, activation='softmax'))\n\n model.compile(loss=loss_f,\n optimizer=opt,\n metrics=['accuracy'])\n print(\"Training new model, loss:\"+loss_f+\", optimizer=\"+opt+\", lstm_len=\"+str(lstm_len)+\", dropoff=\"+str(dropout))\n model.fit(X_train, Y_train, batch_size=16, validation_split = 0.2, epochs=10)\n score = model.evaluate(X_test, Y_test, batch_size=16)\n print(\"\")\n print(\"test score: \" + str(score))\n print(\"\")\n print(\"\")",
"Training new model, loss:categorical_crossentropy, optimizer=sgd, lstm_len=128, dropoff=0.4\nTrain on 32318 samples, validate on 8080 samples\nEpoch 1/10\n32318/32318 [==============================] - 30s 943us/step - loss: 1.0692 - acc: 0.4402 - val_loss: 1.0691 - val_acc: 0.4406\nEpoch 2/10\n32318/32318 [==============================] - 31s 946us/step - loss: 1.0684 - acc: 0.4407 - val_loss: 1.0690 - val_acc: 0.4406\nEpoch 3/10\n32318/32318 [==============================] - 31s 944us/step - loss: 1.0684 - acc: 0.4407 - val_loss: 1.0687 - val_acc: 0.4406\nEpoch 4/10\n32318/32318 [==============================] - 28s 880us/step - loss: 1.0680 - acc: 0.4407 - val_loss: 1.0685 - val_acc: 0.4406\nEpoch 5/10\n32318/32318 [==============================] - 28s 880us/step - loss: 1.0679 - acc: 0.4407 - val_loss: 1.0676 - val_acc: 0.4406\nEpoch 6/10\n32318/32318 [==============================] - 30s 933us/step - loss: 1.0671 - acc: 0.4407 - val_loss: 1.0666 - val_acc: 0.4406\nEpoch 7/10\n32318/32318 [==============================] - 30s 935us/step - loss: 1.0648 - acc: 0.4407 - val_loss: 1.0608 - val_acc: 0.4406\nEpoch 8/10\n32318/32318 [==============================] - 30s 929us/step - loss: 1.0438 - acc: 0.4623 - val_loss: 1.0237 - val_acc: 0.4759\nEpoch 9/10\n32318/32318 [==============================] - 30s 930us/step - loss: 0.9995 - acc: 0.4833 - val_loss: 0.9702 - val_acc: 0.5137\nEpoch 10/10\n32318/32318 [==============================] - 30s 924us/step - loss: 0.9556 - acc: 0.5278 - val_loss: 0.9907 - val_acc: 0.4884\n20122/20122 [==============================] - 5s 251us/step\n\ntest score: [1.0663544713781388, 0.4062220455341625]\n\n\nTraining new model, loss:categorical_crossentropy, optimizer=sgd, lstm_len=128, dropoff=0.45\nTrain on 32318 samples, validate on 8080 samples\nEpoch 1/10\n32318/32318 [==============================] - 35s 1ms/step - loss: 1.0692 - acc: 0.4406 - val_loss: 1.0685 - val_acc: 0.4406\nEpoch 2/10\n32318/32318 [==============================] - 32s 983us/step - loss: 1.0683 - acc: 0.4407 - val_loss: 1.0684 - val_acc: 0.4406\nEpoch 3/10\n32318/32318 [==============================] - 30s 934us/step - loss: 1.0684 - acc: 0.4407 - val_loss: 1.0684 - val_acc: 0.4406\nEpoch 4/10\n32318/32318 [==============================] - 32s 987us/step - loss: 1.0684 - acc: 0.4407 - val_loss: 1.0683 - val_acc: 0.4406\nEpoch 5/10\n32318/32318 [==============================] - 31s 947us/step - loss: 1.0683 - acc: 0.4407 - val_loss: 1.0685 - val_acc: 0.4406\nEpoch 6/10\n32318/32318 [==============================] - 31s 944us/step - loss: 1.0678 - acc: 0.4407 - val_loss: 1.0683 - val_acc: 0.4406\nEpoch 7/10\n32318/32318 [==============================] - 31s 953us/step - loss: 1.0675 - acc: 0.4407 - val_loss: 1.0679 - val_acc: 0.4406\nEpoch 8/10\n32318/32318 [==============================] - 32s 982us/step - loss: 1.0667 - acc: 0.4407 - val_loss: 1.0663 - val_acc: 0.4406\nEpoch 9/10\n32318/32318 [==============================] - 31s 949us/step - loss: 1.0625 - acc: 0.4411 - val_loss: 1.0564 - val_acc: 0.4406\nEpoch 10/10\n32318/32318 [==============================] - 31s 963us/step - loss: 1.0407 - acc: 0.4733 - val_loss: 1.0268 - val_acc: 0.4813\n20122/20122 [==============================] - 5s 262us/step\n\ntest score: [1.02362715051018, 0.49110426399262525]\n\n\nTraining new model, loss:categorical_crossentropy, optimizer=sgd, lstm_len=128, dropoff=0.5\nTrain on 32318 samples, validate on 8080 samples\nEpoch 1/10\n32318/32318 [==============================] - 34s 1ms/step - loss: 1.0695 - acc: 0.4399 - val_loss: 1.0694 - val_acc: 0.4406\nEpoch 2/10\n32318/32318 [==============================] - 31s 969us/step - loss: 1.0688 - acc: 0.4407 - val_loss: 1.0690 - val_acc: 0.4406\nEpoch 3/10\n32318/32318 [==============================] - 31s 957us/step - loss: 1.0685 - acc: 0.4407 - val_loss: 1.0686 - val_acc: 0.4406\nEpoch 4/10\n32318/32318 [==============================] - 32s 986us/step - loss: 1.0684 - acc: 0.4407 - val_loss: 1.0684 - val_acc: 0.4406\nEpoch 5/10\n32318/32318 [==============================] - 32s 987us/step - loss: 1.0684 - acc: 0.4407 - val_loss: 1.0684 - val_acc: 0.4406\nEpoch 6/10\n32318/32318 [==============================] - 32s 991us/step - loss: 1.0684 - acc: 0.4407 - val_loss: 1.0683 - val_acc: 0.4406\nEpoch 7/10\n32318/32318 [==============================] - 31s 963us/step - loss: 1.0683 - acc: 0.4407 - val_loss: 1.0683 - val_acc: 0.4406\nEpoch 8/10\n32318/32318 [==============================] - 31s 962us/step - loss: 1.0683 - acc: 0.4407 - val_loss: 1.0682 - val_acc: 0.4406\nEpoch 9/10\n32318/32318 [==============================] - 32s 991us/step - loss: 1.0680 - acc: 0.4407 - val_loss: 1.0678 - val_acc: 0.4406\nEpoch 10/10\n32318/32318 [==============================] - 33s 1ms/step - loss: 1.0675 - acc: 0.4407 - val_loss: 1.0673 - val_acc: 0.4406\n20122/20122 [==============================] - 6s 274us/step\n\ntest score: [1.0238210319844738, 0.5285756883043239]\n\n\nTraining new model, loss:categorical_crossentropy, optimizer=sgd, lstm_len=128, dropoff=0.55\nTrain on 32318 samples, validate on 8080 samples\nEpoch 1/10\n32318/32318 [==============================] - 35s 1ms/step - loss: 1.0692 - acc: 0.4406 - val_loss: 1.0684 - val_acc: 0.4406\nEpoch 2/10\n32318/32318 [==============================] - 33s 1ms/step - loss: 1.0687 - acc: 0.4407 - val_loss: 1.0687 - val_acc: 0.4406\nEpoch 3/10\n32318/32318 [==============================] - 33s 1ms/step - loss: 1.0684 - acc: 0.4407 - val_loss: 1.0682 - val_acc: 0.4406\nEpoch 4/10\n32318/32318 [==============================] - 32s 991us/step - loss: 1.0683 - acc: 0.4407 - val_loss: 1.0682 - val_acc: 0.4406\nEpoch 5/10\n32318/32318 [==============================] - 32s 978us/step - loss: 1.0682 - acc: 0.4407 - val_loss: 1.0678 - val_acc: 0.4406\nEpoch 6/10\n32318/32318 [==============================] - 32s 999us/step - loss: 1.0676 - acc: 0.4407 - val_loss: 1.0689 - val_acc: 0.4406\nEpoch 7/10\n32318/32318 [==============================] - 32s 999us/step - loss: 1.0672 - acc: 0.4407 - val_loss: 1.0665 - val_acc: 0.4406\nEpoch 8/10\n32318/32318 [==============================] - 32s 999us/step - loss: 1.0652 - acc: 0.4408 - val_loss: 1.0623 - val_acc: 0.4406\nEpoch 9/10\n32318/32318 [==============================] - 32s 1ms/step - loss: 1.0509 - acc: 0.4624 - val_loss: 1.0352 - val_acc: 0.4847\nEpoch 10/10\n32318/32318 [==============================] - 33s 1ms/step - loss: 1.0279 - acc: 0.4883 - val_loss: 1.0159 - val_acc: 0.4948\n20122/20122 [==============================] - 6s 300us/step\n\ntest score: [1.0234103390857934, 0.49726667329587537]\n\n\nTraining new model, loss:categorical_crossentropy, optimizer=sgd, lstm_len=128, dropoff=0.6\nTrain on 32318 samples, validate on 8080 samples\nEpoch 1/10\n32318/32318 [==============================] - 38s 1ms/step - loss: 1.0694 - acc: 0.4406 - val_loss: 1.0685 - val_acc: 0.4406\nEpoch 2/10\n32318/32318 [==============================] - 33s 1ms/step - loss: 1.0684 - acc: 0.4407 - val_loss: 1.0686 - val_acc: 0.4406\nEpoch 3/10\n32318/32318 [==============================] - 34s 1ms/step - loss: 1.0685 - acc: 0.4407 - val_loss: 1.0696 - val_acc: 0.4406\nEpoch 4/10\n32318/32318 [==============================] - 35s 1ms/step - loss: 1.0680 - acc: 0.4407 - val_loss: 1.0685 - val_acc: 0.4406\nEpoch 5/10\n32318/32318 [==============================] - 34s 1ms/step - loss: 1.0672 - acc: 0.4407 - val_loss: 1.0664 - val_acc: 0.4406\nEpoch 6/10\n32318/32318 [==============================] - 34s 1ms/step - loss: 1.0639 - acc: 0.4407 - val_loss: 1.0578 - val_acc: 0.4406\nEpoch 7/10\n32318/32318 [==============================] - 33s 1ms/step - loss: 1.0414 - acc: 0.4698 - val_loss: 1.0244 - val_acc: 0.4806\nEpoch 8/10\n32318/32318 [==============================] - 33s 1ms/step - loss: 1.0036 - acc: 0.4833 - val_loss: 0.9859 - val_acc: 0.5181\nEpoch 9/10\n32318/32318 [==============================] - 33s 1ms/step - loss: 0.9609 - acc: 0.5228 - val_loss: 0.9430 - val_acc: 0.5547\nEpoch 10/10\n32318/32318 [==============================] - 33s 1ms/step - loss: 0.9401 - acc: 0.5384 - val_loss: 0.9377 - val_acc: 0.5335\n20122/20122 [==============================] - 6s 285us/step\n\ntest score: [1.0087274505276647, 0.5294205347499462]\n\n\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d000adb69592d40c5cafd8a3c20112357e08b63f | 295,491 | ipynb | Jupyter Notebook | parse_results_with_visualization/Hyper_params_visualization.ipynb | HenryNebula/Personalization_Final_Project | 5d18a8628bed2dfd2894b9d2f33c1e9a5df27ecc | [
"MIT"
] | 1 | 2020-11-03T18:02:15.000Z | 2020-11-03T18:02:15.000Z | parse_results_with_visualization/Hyper_params_visualization.ipynb | HenryNebula/Personalization_Final_Project | 5d18a8628bed2dfd2894b9d2f33c1e9a5df27ecc | [
"MIT"
] | null | null | null | parse_results_with_visualization/Hyper_params_visualization.ipynb | HenryNebula/Personalization_Final_Project | 5d18a8628bed2dfd2894b9d2f33c1e9a5df27ecc | [
"MIT"
] | 1 | 2020-03-22T01:01:21.000Z | 2020-03-22T01:01:21.000Z | 286.884466 | 40,604 | 0.920891 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib\nimport seaborn as sns\nimport matplotlib.pyplot as plt\npd.set_option('display.max_colwidth', -1)",
"_____no_output_____"
],
[
"default = pd.read_csv('./results/results_default.csv')",
"_____no_output_____"
],
[
"new = pd.read_csv('./results/results_new.csv')",
"_____no_output_____"
],
[
"selected_cols = ['model','hyper','metric','value']",
"_____no_output_____"
],
[
"default = default[selected_cols]",
"_____no_output_____"
],
[
"new = new[selected_cols]",
"_____no_output_____"
],
[
"default.model.unique()",
"_____no_output_____"
],
[
"#function to extract nested info\ndef split_params(df):\n join_table = df.copy()\n join_table[\"list_hyper\"] = join_table[\"hyper\"].apply(eval)\n join_table = join_table.explode(\"list_hyper\")\n join_table[\"params_name\"], join_table[\"params_val\"] = zip(*join_table[\"list_hyper\"])\n\n return join_table\n",
"_____no_output_____"
],
[
"color = ['lightpink','skyblue','lightgreen', \"lightgrey\", \"navajowhite\", \"thistle\"]\nmarkerfacecolor = ['red', 'blue', 'green','grey', \"orangered\", \"darkviolet\" ]\nmarker = ['P', '^' ,'o', \"H\", \"X\", \"p\"]\nfig_size=(6,4)",
"_____no_output_____"
]
],
[
[
"### Default server",
"_____no_output_____"
]
],
[
[
"default_split = split_params(default)[['model','metric','value','params_name','params_val']]",
"_____no_output_____"
],
[
"models = default_split.model.unique().tolist()",
"_____no_output_____"
],
[
"CollectiveMF_Item_set = default_split[default_split['model'] == models[0]]\nCollectiveMF_User_set = default_split[default_split['model'] == models[1]]\nCollectiveMF_No_set = default_split[default_split['model'] == models[2]]\nCollectiveMF_Both_set = default_split[default_split['model'] == models[3]]\nsurprise_SVD_set = default_split[default_split['model'] == models[4]]\nsurprise_Baseline_set = default_split[default_split['model'] == models[5]]",
"_____no_output_____"
]
],
[
[
"## surprise_SVD",
"_____no_output_____"
]
],
[
[
"surprise_SVD_ndcg = surprise_SVD_set[(surprise_SVD_set['metric'] == 'ndcg@10')]\nsurprise_SVD_ndcg = surprise_SVD_ndcg.pivot(index= 'value', \n columns='params_name', \n values='params_val').reset_index(inplace = False)",
"_____no_output_____"
],
[
"surprise_SVD_ndcg = surprise_SVD_ndcg[surprise_SVD_ndcg.n_factors > 4]",
"_____no_output_____"
],
[
"n_factors = [10,50,100,150]\nreg_all = [0.01,0.05,0.1,0.5]\nlr_all = [0.002,0.005,0.01]\n",
"_____no_output_____"
],
[
"surprise_SVD_ndcg = surprise_SVD_ndcg.sort_values('reg_all')\nfig, ax = plt.subplots(1,1, figsize = fig_size)\nfor i in range(4):\n labelstring = 'n_factors = '+ str(n_factors[i])\n ax.semilogx('reg_all', 'value', \n data = surprise_SVD_ndcg.loc[(surprise_SVD_ndcg['lr_all'] == 0.002)&(surprise_SVD_ndcg['n_factors']== n_factors[i])],\n marker= marker[i], markerfacecolor=markerfacecolor[i], markersize=9,\n color= color[i], linewidth=3, label = labelstring)\nax.legend()\nax.set_ylabel('ndcg@10',fontsize = 18)\nax.set_xlabel('regParam',fontsize = 18)\nax.set_title('surprise_SVD \\n ndcg@10 vs regParam with lr = 0.002',fontsize = 18)\nax.set_xticks(reg_all)\nax.xaxis.set_tick_params(labelsize=14)\nax.yaxis.set_tick_params(labelsize=13)\npic = fig\nplt.tight_layout()\n",
"_____no_output_____"
],
[
"pic.savefig('figs/hyper/SVD_ndcg_vs_reg_factor.eps', format='eps')",
"The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\nThe PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\n"
],
[
"surprise_SVD_ndcg = surprise_SVD_ndcg.sort_values('n_factors')\nfig, ax = plt.subplots(1,1, figsize = fig_size)\nfor i in range(4):\n labelstring = 'regParam = '+ str(reg_all[i])\n ax.plot('n_factors', 'value', \n data = surprise_SVD_ndcg.loc[(surprise_SVD_ndcg['lr_all'] == 0.002)&(surprise_SVD_ndcg['reg_all']== reg_all[i])],\n marker= marker[i], markerfacecolor=markerfacecolor[i], markersize=9,\n color= color[i], linewidth=3, label = labelstring)\nax.legend()\nax.set_ylabel('ndcg@10',fontsize = 18)\nax.set_xlabel('n_factors',fontsize = 18)\nax.set_title('surprise_SVD \\n ndcg@10 vs n_factors with lr = 0.002',fontsize = 18)\nax.set_xticks(n_factors)\nax.xaxis.set_tick_params(labelsize=14)\nax.yaxis.set_tick_params(labelsize=13)\n\npic = fig\nplt.tight_layout()",
"_____no_output_____"
],
[
"pic.savefig('figs/hyper/SVD_ndcg_vs_factor_reg.eps', format='eps')",
"The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\nThe PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\n"
]
],
[
[
"## CollectiveMF_Both",
"_____no_output_____"
]
],
[
[
"reg_param = [0.0001, 0.001, 0.01]\nw_main = [0.5, 0.6, 0.7, 0.8, 0.9, 1.0]\nk = [4.,8.,16.]\n",
"_____no_output_____"
],
[
"CollectiveMF_Both_ndcg = CollectiveMF_Both_set[CollectiveMF_Both_set['metric'] == 'ndcg@10']",
"_____no_output_____"
],
[
"CollectiveMF_Both_ndcg = CollectiveMF_Both_ndcg.pivot(index= 'value', \n columns='params_name', \n values='params_val').reset_index(inplace = False)",
"_____no_output_____"
],
[
"### Visualization of hyperparameters tuning\n\nfig, ax = plt.subplots(1,1, figsize = fig_size)\nCollectiveMF_Both_ndcg.sort_values(\"reg_param\", inplace=True)\nfor i in range(len(w_main)):\n labelstring = 'w_main = '+ str(w_main[i])\n ax.semilogx('reg_param', 'value', \n data = CollectiveMF_Both_ndcg.loc[(CollectiveMF_Both_ndcg['k'] == 4.0)&(CollectiveMF_Both_ndcg['w_main']== w_main[i])],\n marker= marker[i], markerfacecolor= markerfacecolor[i], markersize=9,\n color= color[i], linewidth=3, label = labelstring)\n\nax.legend()\nax.set_ylabel('ndcg@10',fontsize = 18)\nax.set_xlabel('regParam',fontsize = 18)\nax.set_title('CollectiveMF_Both \\n ndcg@10 vs regParam with k = 4.0',fontsize = 18)\nax.set_xticks(reg_param)\nax.xaxis.set_tick_params(labelsize=10)\nax.yaxis.set_tick_params(labelsize=13)\npic = fig\nplt.tight_layout()",
"_____no_output_____"
],
[
"pic.savefig('figs/hyper/CMF_ndcg_vs_reg_w_main.eps', format='eps')",
"The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\nThe PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\n"
],
[
"fig, ax = plt.subplots(1,1, figsize = fig_size)\nCollectiveMF_Both_ndcg = CollectiveMF_Both_ndcg.sort_values('w_main')\nfor i in range(len(reg_param)):\n labelstring = 'regParam = '+ str(reg_param[i])\n ax.plot('w_main', 'value', \n data = CollectiveMF_Both_ndcg.loc[(CollectiveMF_Both_ndcg['k'] == 4.0)&(CollectiveMF_Both_ndcg['reg_param']== reg_param[i])],\n marker= marker[i], markerfacecolor= markerfacecolor[i], markersize=9,\n color= color[i], linewidth=3, label = labelstring)\n\nax.legend()\nax.set_ylabel('ndcg@10',fontsize = 18)\nax.set_xlabel('w_main',fontsize = 18)\nax.set_title('CollectiveMF_Both \\n ndcg@10 vs w_main with k = 4.0',fontsize = 18)\nax.set_xticks(w_main)\nax.xaxis.set_tick_params(labelsize=14)\nax.yaxis.set_tick_params(labelsize=13)\npic = fig\nplt.tight_layout()",
"_____no_output_____"
],
[
"pic.savefig('figs/hyper/CMF_ndcg_vs_w_main_reg.eps', format='eps')",
"The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\nThe PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\n"
]
],
[
[
"### New server",
"_____no_output_____"
]
],
[
[
"new_split = split_params(new)[['model','metric','value','params_name','params_val']]",
"_____no_output_____"
],
[
"Test_implicit_set = new_split[new_split['model'] == 'BPR']\nFMItem_set = new_split[new_split['model'] == 'FMItem']\nFMNone_set = new_split[new_split['model'] == 'FMNone']",
"_____no_output_____"
]
],
[
[
"## Test_implicit",
"_____no_output_____"
]
],
[
[
"Test_implicit_set_ndcg = Test_implicit_set[Test_implicit_set['metric'] == 'ndcg@10']",
"_____no_output_____"
],
[
"Test_implicit_set_ndcg = Test_implicit_set_ndcg.pivot(index=\"value\", \n columns='params_name', \n values='params_val').reset_index(inplace = False)",
"_____no_output_____"
],
[
"Test_implicit_set_ndcg = Test_implicit_set_ndcg[Test_implicit_set_ndcg.iteration > 20].copy()",
"_____no_output_____"
],
[
"regularization = [0.001,0.005, 0.01 ]\nlearning_rate = [0.0001, 0.001, 0.005]\nfactors = [4,8,16]",
"_____no_output_____"
],
[
"Test_implicit_set_ndcg.sort_values('regularization', inplace=True)\nfig, ax = plt.subplots(1,1, figsize = fig_size)\nfor i in range(len(factors)):\n labelstring = 'n_factors = '+ str(factors[i])\n ax.plot('regularization', 'value', \n data = Test_implicit_set_ndcg.loc[(Test_implicit_set_ndcg['learning_rate'] == 0.005)&(Test_implicit_set_ndcg['factors']== factors[i])],\n marker= marker[i], markerfacecolor=markerfacecolor[i], markersize=9,\n color= color[i], linewidth=3, label = labelstring)\nax.legend() \nax.set_ylabel('ndcg@10',fontsize = 18)\nax.set_xlabel('regParam',fontsize = 18)\nax.set_title('BPR \\n ndcg@10 vs regParam with lr = 0.005',fontsize = 18)\nax.set_xticks([1e-3, 5e-3, 1e-2])\nax.xaxis.set_tick_params(labelsize=14)\nax.yaxis.set_tick_params(labelsize=13)\npic = fig\nplt.tight_layout()",
"_____no_output_____"
],
[
"pic.savefig('figs/hyper/BPR_ndcg_vs_reg_factors.eps', format='eps')",
"The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\nThe PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\n"
],
[
"Test_implicit_set_ndcg.sort_values('factors', inplace=True)\nfig, ax = plt.subplots(1,1, figsize = fig_size)\nfor i in range(len(regularization)):\n labelstring = 'regParam = '+ str(regularization[i])\n ax.plot('factors', 'value', \n data = Test_implicit_set_ndcg.loc[(Test_implicit_set_ndcg['learning_rate'] == 0.005)&\n (Test_implicit_set_ndcg.regularization== regularization[i])],\n marker= marker[i], markerfacecolor=markerfacecolor[i], markersize=9,\n color= color[i], linewidth=3, label = labelstring)\nax.legend() \nax.set_ylabel('ndcg@10',fontsize = 18)\nax.set_xlabel('n_factors',fontsize = 18)\nax.set_title('BPR \\n ndcg@10 vs n_factors with lr = 0.005',fontsize = 18)\nax.set_xticks(factors)\nax.xaxis.set_tick_params(labelsize=14)\nax.yaxis.set_tick_params(labelsize=13)\npic = fig\nplt.tight_layout()",
"_____no_output_____"
],
[
"pic.savefig('figs/hyper/BPR_ndcg_vs_factors_reg.eps', format='eps',fontsize = 18)",
"The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\nThe PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\n"
]
],
[
[
"## FMItem",
"_____no_output_____"
]
],
[
[
"FMItem_set_ndcg = FMItem_set[FMItem_set['metric'] == 'ndcg@10']",
"_____no_output_____"
],
[
"FMItem_set_ndcg = FMItem_set_ndcg.pivot(index=\"value\", \n columns='params_name', \n values='params_val').reset_index(inplace = False)",
"_____no_output_____"
],
[
"FMItem_set_ndcg = FMItem_set_ndcg[(FMItem_set_ndcg.n_iter == 100) & (FMItem_set_ndcg[\"rank\"] <= 4)].copy()",
"_____no_output_____"
],
[
"FMItem_set_ndcg",
"_____no_output_____"
],
[
"color = ['lightpink','skyblue','lightgreen', \"lightgrey\", \"navajowhite\", \"thistle\"]\nmarkerfacecolor = ['red', 'blue', 'green','grey', \"orangered\", \"darkviolet\" ]\nmarker = ['P', '^' ,'o', \"H\", \"X\", \"p\"]\nreg = [0.2, 0.3, 0.5, 0.8, 0.9, 1]\nfct = [2,4]",
"_____no_output_____"
],
[
"FMItem_set_ndcg.sort_values('l2_reg_V', inplace=True)\nfig, ax = plt.subplots(1,1, figsize = fig_size)\nfor i in range(len(reg)):\n labelstring = 'regParam = '+ str(reg[i])\n ax.plot('rank', 'value', \n data = FMItem_set_ndcg.loc[(FMItem_set_ndcg.l2_reg_V == reg[i])&\n (FMItem_set_ndcg.l2_reg_w == reg[i])],\n marker= marker[i], markerfacecolor=markerfacecolor[i], markersize=9,\n color= color[i], linewidth=3, label = labelstring)\nax.legend() \nax.set_ylabel('ndcg@10',fontsize = 18)\nax.set_xlabel('n_factors',fontsize = 18)\nax.set_title('FM_Item \\n ndcg@10 vs n_factors with lr = 0.005',fontsize = 18)\nax.set_xticks(fct)\nax.xaxis.set_tick_params(labelsize=14)\nax.yaxis.set_tick_params(labelsize=13)\npic = fig\nplt.tight_layout()",
"_____no_output_____"
],
[
"pic.savefig('figs/hyper/FM_ndcg_vs_factors_reg.eps', format='eps',fontsize = 18)",
"The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\nThe PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\n"
],
[
"FMItem_set_ndcg.sort_values('rank', inplace=True)\nfig, ax = plt.subplots(1,1, figsize = fig_size)\nfor i in range(len(fct)):\n labelstring = 'n_factors = '+ str(fct[i])\n ax.plot('l2_reg_V', 'value', \n data = FMItem_set_ndcg.loc[(FMItem_set_ndcg[\"rank\"] == fct[i])],\n marker= marker[i], markerfacecolor=markerfacecolor[i], markersize=9,\n color= color[i], linewidth=3, label = labelstring)\nax.legend() \nax.set_ylabel('ndcg@10',fontsize = 18)\nax.set_xlabel('regParam',fontsize = 18)\nax.set_title('FM_Item \\n ndcg@10 vs n_factors with lr = 0.005',fontsize = 18)\nax.set_xticks(np.arange(0.1, 1.1, 0.1))\nax.xaxis.set_tick_params(labelsize=14)\nax.yaxis.set_tick_params(labelsize=13)\npic = fig\nplt.tight_layout()",
"_____no_output_____"
],
[
"pic.savefig('figs/hyper/FM_ndcg_vs_reg_factors.eps', format='eps')",
"The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\nThe PostScript backend does not support transparency; partially transparent artists will be rendered opaque.\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d000b22fa28b30bab7e79443b600a0d64a9732ce | 5,071 | ipynb | Jupyter Notebook | import_file_checker_result.ipynb | acdh-oeaw/4dpuzzle | 7856bbd82c7dfa8da1d5f1ad40593219a35b3cfe | [
"MIT"
] | null | null | null | import_file_checker_result.ipynb | acdh-oeaw/4dpuzzle | 7856bbd82c7dfa8da1d5f1ad40593219a35b3cfe | [
"MIT"
] | 6 | 2020-06-05T18:32:02.000Z | 2022-02-10T07:22:24.000Z | import_file_checker_result.ipynb | acdh-oeaw/4dpuzzle | 7856bbd82c7dfa8da1d5f1ad40593219a35b3cfe | [
"MIT"
] | 1 | 2020-06-30T13:52:41.000Z | 2020-06-30T13:52:41.000Z | 22.842342 | 128 | 0.521199 | [
[
[
"import json\nimport pandas as pd",
"_____no_output_____"
],
[
"from webpage.appcreator.import_utils import fetch_models\nfrom filechecker.filechecker_utils import filechecker_to_df, find_matching_objects",
"_____no_output_____"
],
[
"# file = r\"/mnt/OeAW_Projekte03/OREA-EGYPT_Puzzle4D/ARCHE-preparation/checkReports/2019_11_26_Puzzle4d/fileList.json\"\n# file = r\"/home/csae8092/repos/p4d/data/fileList.json\"\nfile = r\"/home/csae8092/repos/p4d/fileList.json\" # add missing dbs 2020-12-03",
"_____no_output_____"
],
[
"# data = json.load(open(file, \"r\", encoding=\"utf-8\"))",
"_____no_output_____"
],
[
"# df = filechecker_to_df(file)\nmain_df = filechecker_to_df(file)",
"_____no_output_____"
],
[
"df = main_df.loc[main_df['directory'].str.contains(\"Datenban\")]",
"_____no_output_____"
],
[
"# all_models = fetch_models('archiv')\nall_models = [x for x in fetch_models('archiv')if 'Datenbase' in x.__name__] # add missing dbs 2020-12-03",
"_____no_output_____"
],
[
"all_models",
"_____no_output_____"
],
[
"for i, row in df.iterrows():\n leg_id = row['filename'].lower()\n leg_id = leg_id.split('.')[0]\n my_obj = find_matching_objects(all_models, leg_id)\n if my_obj is None:\n continue\n else:\n for prop in row.index:\n my_prop = f\"fc_{prop}\"\n my_val = f\"{row[prop]}\"\n setattr(my_obj, my_prop, my_val)\n my_obj.fc_match = True\n my_obj.save()",
"_____no_output_____"
],
[
"items = Inventorybooks.objects.all()",
"_____no_output_____"
],
[
"items.count()",
"_____no_output_____"
],
[
"items.filter(fc_match=True).count()",
"_____no_output_____"
],
[
"def fc_match_quota(app_name):\n summary = [\n [\"class_name\", 'all_objects', 'matching_resource', 'no_matching_resource', 'percentage']\n ]\n all_models = fetch_models(app_name)\n for x in all_models:\n items = x.objects.all()\n all_objects = items.count()\n matches = x.objects.filter(fc_match=True).count()\n misses = x.objects.exclude(fc_match=True).count()\n class_name = x.__name__\n try:\n percentage = (matches / all_objects)*100 \n except ZeroDivisionError:\n percentage = 0\n summary.append(\n [class_name, all_objects, matches, misses, percentage]\n )\n return summary",
"_____no_output_____"
],
[
"table = fc_match_quota('archiv')",
"_____no_output_____"
],
[
"df = pd.DataFrame(table, columns=table[0])",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.to_csv('matching_binaries.csv')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d000b6b92fe17366a6da83e58577a3929840b135 | 585,062 | ipynb | Jupyter Notebook | src/VotingClassifier/.ipynb_checkpoints/knn-checkpoint.ipynb | joaquinfontela/Machine-Learning | 733284fe82e6c128358fe2e7721d887e2683da9f | [
"MIT"
] | null | null | null | src/VotingClassifier/.ipynb_checkpoints/knn-checkpoint.ipynb | joaquinfontela/Machine-Learning | 733284fe82e6c128358fe2e7721d887e2683da9f | [
"MIT"
] | null | null | null | src/VotingClassifier/.ipynb_checkpoints/knn-checkpoint.ipynb | joaquinfontela/Machine-Learning | 733284fe82e6c128358fe2e7721d887e2683da9f | [
"MIT"
] | 1 | 2021-07-30T20:53:53.000Z | 2021-07-30T20:53:53.000Z | 62.04263 | 1,432 | 0.546662 | [
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nfrom matplotlib import style\nimport matplotlib.ticker as ticker\nimport seaborn as sns",
"_____no_output_____"
],
[
"from sklearn.datasets import load_boston\nfrom sklearn.ensemble import RandomForestClassifier, VotingClassifier, GradientBoostingClassifier\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import plot_confusion_matrix\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import f1_score, make_scorer\nfrom sklearn.compose import ColumnTransformer\nfrom sklearn.preprocessing import OneHotEncoder\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.model_selection import RepeatedKFold\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.model_selection import ParameterGrid\nfrom sklearn.inspection import permutation_importance\nimport multiprocessing",
"_____no_output_____"
],
[
"from xgboost import XGBClassifier",
"_____no_output_____"
],
[
"labels = pd.read_csv('../../csv/train_labels.csv')\nlabels.head()",
"_____no_output_____"
],
[
"values = pd.read_csv('../../csv/train_values.csv')\nvalues.T",
"_____no_output_____"
],
[
"to_be_categorized = [\"land_surface_condition\", \"foundation_type\", \"roof_type\",\\\n \"position\", \"ground_floor_type\", \"other_floor_type\",\\\n \"plan_configuration\", \"legal_ownership_status\"]\nfor row in to_be_categorized:\n values[row] = values[row].astype(\"category\")\nvalues.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 260601 entries, 0 to 260600\nData columns (total 39 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 building_id 260601 non-null int64 \n 1 geo_level_1_id 260601 non-null int64 \n 2 geo_level_2_id 260601 non-null int64 \n 3 geo_level_3_id 260601 non-null int64 \n 4 count_floors_pre_eq 260601 non-null int64 \n 5 age 260601 non-null int64 \n 6 area_percentage 260601 non-null int64 \n 7 height_percentage 260601 non-null int64 \n 8 land_surface_condition 260601 non-null category\n 9 foundation_type 260601 non-null category\n 10 roof_type 260601 non-null category\n 11 ground_floor_type 260601 non-null category\n 12 other_floor_type 260601 non-null category\n 13 position 260601 non-null category\n 14 plan_configuration 260601 non-null category\n 15 has_superstructure_adobe_mud 260601 non-null int64 \n 16 has_superstructure_mud_mortar_stone 260601 non-null int64 \n 17 has_superstructure_stone_flag 260601 non-null int64 \n 18 has_superstructure_cement_mortar_stone 260601 non-null int64 \n 19 has_superstructure_mud_mortar_brick 260601 non-null int64 \n 20 has_superstructure_cement_mortar_brick 260601 non-null int64 \n 21 has_superstructure_timber 260601 non-null int64 \n 22 has_superstructure_bamboo 260601 non-null int64 \n 23 has_superstructure_rc_non_engineered 260601 non-null int64 \n 24 has_superstructure_rc_engineered 260601 non-null int64 \n 25 has_superstructure_other 260601 non-null int64 \n 26 legal_ownership_status 260601 non-null category\n 27 count_families 260601 non-null int64 \n 28 has_secondary_use 260601 non-null int64 \n 29 has_secondary_use_agriculture 260601 non-null int64 \n 30 has_secondary_use_hotel 260601 non-null int64 \n 31 has_secondary_use_rental 260601 non-null int64 \n 32 has_secondary_use_institution 260601 non-null int64 \n 33 has_secondary_use_school 260601 non-null int64 \n 34 has_secondary_use_industry 260601 non-null int64 \n 35 has_secondary_use_health_post 260601 non-null int64 \n 36 has_secondary_use_gov_office 260601 non-null int64 \n 37 has_secondary_use_use_police 260601 non-null int64 \n 38 has_secondary_use_other 260601 non-null int64 \ndtypes: category(8), int64(31)\nmemory usage: 63.6 MB\n"
],
[
"datatypes = dict(values.dtypes)\nfor row in values.columns:\n if datatypes[row] != \"int64\" and datatypes[row] != \"int32\" and \\\n datatypes[row] != \"int16\" and datatypes[row] != \"int8\":\n continue\n if values[row].nlargest(1).item() > 32767 and values[row].nlargest(1).item() < 2**31:\n values[row] = values[row].astype(np.int32)\n elif values[row].nlargest(1).item() > 127:\n values[row] = values[row].astype(np.int16)\n else:\n values[row] = values[row].astype(np.int8)",
"_____no_output_____"
],
[
"labels[\"building_id\"] = labels[\"building_id\"].astype(np.int32)\nlabels[\"damage_grade\"] = labels[\"damage_grade\"].astype(np.int8)\nlabels.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 260601 entries, 0 to 260600\nData columns (total 2 columns):\n # Column Non-Null Count Dtype\n--- ------ -------------- -----\n 0 building_id 260601 non-null int32\n 1 damage_grade 260601 non-null int8 \ndtypes: int32(1), int8(1)\nmemory usage: 1.2 MB\n"
]
],
[
[
"# Feature Engineering para XGBoost",
"_____no_output_____"
]
],
[
[
"important_values = values\\\n .merge(labels, on=\"building_id\")\nimportant_values.drop(columns=[\"building_id\"], inplace = True)\nimportant_values[\"geo_level_1_id\"] = important_values[\"geo_level_1_id\"].astype(\"category\")\nimportant_values",
"_____no_output_____"
],
[
"\nX_train, X_test, y_train, y_test = train_test_split(important_values.drop(columns = 'damage_grade'),\n important_values['damage_grade'], test_size = 0.2, random_state = 123)",
"_____no_output_____"
],
[
"#OneHotEncoding\ndef encode_and_bind(original_dataframe, feature_to_encode):\n dummies = pd.get_dummies(original_dataframe[[feature_to_encode]])\n res = pd.concat([original_dataframe, dummies], axis=1)\n res = res.drop([feature_to_encode], axis=1)\n return(res) \n\nfeatures_to_encode = [\"geo_level_1_id\", \"land_surface_condition\", \"foundation_type\", \"roof_type\",\\\n \"position\", \"ground_floor_type\", \"other_floor_type\",\\\n \"plan_configuration\", \"legal_ownership_status\"]\nfor feature in features_to_encode:\n X_train = encode_and_bind(X_train, feature)\n X_test = encode_and_bind(X_test, feature)",
"_____no_output_____"
],
[
"X_train",
"_____no_output_____"
],
[
"import time\n\n# min_child_weight = [0, 1, 2]\n# max_delta_step = [0, 5, 10]\n\ndef my_grid_search():\n print(time.gmtime())\n i = 1\n df = pd.DataFrame({'subsample': [],\n 'gamma': [],\n 'learning_rate': [],\n 'max_depth': [],\n 'score': []})\n for subsample in [0.75, 0.885, 0.95]:\n for gamma in [0.75, 1, 1.25]:\n for learning_rate in [0.4375, 0.45, 0.4625]:\n for max_depth in [5, 6, 7]:\n model = XGBClassifier(n_estimators = 350,\n booster = 'gbtree',\n subsample = subsample,\n gamma = gamma,\n max_depth = max_depth,\n learning_rate = learning_rate,\n label_encoder = False,\n verbosity = 0)\n model.fit(X_train, y_train)\n y_preds = model.predict(X_test)\n score = f1_score(y_test, y_preds, average = 'micro')\n df = df.append(pd.Series(\n data={'subsample': subsample,\n 'gamma': gamma,\n 'learning_rate': learning_rate,\n 'max_depth': max_depth,\n 'score': score},\n name = i))\n print(i, time.gmtime())\n i += 1\n\n return df.sort_values('score', ascending = False)\n\ncurrent_df = my_grid_search()\ndf = pd.read_csv('grid-search/res-feature-engineering.csv')\ndf.append(current_df)\ndf.to_csv('grid-search/res-feature-engineering.csv')\n\ncurrent_df",
"time.struct_time(tm_year=2021, tm_mon=7, tm_mday=13, tm_hour=5, tm_min=48, tm_sec=50, tm_wday=1, tm_yday=194, tm_isdst=0)\n"
],
[
"import time\n\ndef my_grid_search():\n print(time.gmtime())\n i = 1\n df = pd.DataFrame({'subsample': [],\n 'gamma': [],\n 'learning_rate': [],\n 'max_depth': [],\n 'score': []})\n for subsample in [0.885]:\n for gamma in [1]:\n for learning_rate in [0.45]:\n for max_depth in [5,6,7,8]:\n model = XGBClassifier(n_estimators = 350,\n booster = 'gbtree',\n subsample = subsample,\n gamma = gamma,\n max_depth = max_depth,\n learning_rate = learning_rate,\n label_encoder = False,\n verbosity = 0)\n model.fit(X_train, y_train)\n y_preds = model.predict(X_test)\n score = f1_score(y_test, y_preds, average = 'micro')\n df = df.append(pd.Series(\n data={'subsample': subsample,\n 'gamma': gamma,\n 'learning_rate': learning_rate,\n 'max_depth': max_depth,\n 'score': score},\n name = i))\n print(i, time.gmtime())\n i += 1\n\n return df.sort_values('score', ascending = False)\n\ndf = my_grid_search()\n# df = pd.read_csv('grid-search/res-feature-engineering.csv')\n# df.append(current_df)\ndf.to_csv('grid-search/res-feature-engineering.csv')\n\ndf",
"time.struct_time(tm_year=2021, tm_mon=7, tm_mday=13, tm_hour=21, tm_min=22, tm_sec=10, tm_wday=1, tm_yday=194, tm_isdst=0)\n"
],
[
"pd.read_csv('grid-search/res-no-feature-engineering.csv')\\\n .nlargest(20, 'score')",
"_____no_output_____"
]
],
[
[
"# Entreno tres de los mejores modelos con Voting.",
"_____no_output_____"
]
],
[
[
"xgb_model_1 = XGBClassifier(n_estimators = 350,\n subsample = 0.885,\n booster = 'gbtree',\n gamma = 1,\n learning_rate = 0.45,\n label_encoder = False,\n verbosity = 2)\n\nxgb_model_2 = XGBClassifier(n_estimators = 350,\n subsample = 0.950,\n booster = 'gbtree',\n gamma = 0.5,\n learning_rate = 0.45,\n label_encoder = False,\n verbosity = 2)\n\nxgb_model_3 = XGBClassifier(n_estimators = 350,\n subsample = 0.750,\n booster = 'gbtree',\n gamma = 1,\n learning_rate = 0.45,\n label_encoder = False,\n verbosity = 2)\n\nxgb_model_4 = XGBClassifier(n_estimators = 350,\n subsample = 0.80,\n booster = 'gbtree',\n gamma = 1,\n learning_rate = 0.55,\n label_encoder = False,\n verbosity = 2)",
"_____no_output_____"
],
[
"rf_model_1 = RandomForestClassifier(n_estimators = 150,\n max_depth = None,\n max_features = 45,\n min_samples_split = 15,\n min_samples_leaf = 1,\n criterion = \"gini\",\n verbose=True)\n\nrf_model_2 = RandomForestClassifier(n_estimators = 250,\n max_depth = None,\n max_features = 45,\n min_samples_split = 15,\n min_samples_leaf = 1,\n criterion = \"gini\",\n verbose=True,\n n_jobs =-1)",
"_____no_output_____"
],
[
"import lightgbm as lgb\nlgbm_model_1 = lgb.LGBMClassifier(boosting_type='gbdt', \n colsample_bytree=1.0,\n importance_type='split', \n learning_rate=0.15,\n max_depth=None,\n n_estimators=1600, \n n_jobs=-1,\n objective=None,\n subsample=1.0, \n subsample_for_bin=200000, \n subsample_freq=0)\n\nlgbm_model_2 = lgb.LGBMClassifier(boosting_type='gbdt', \n colsample_bytree=1.0,\n importance_type='split', \n learning_rate=0.15,\n max_depth=25,\n n_estimators=1750, \n n_jobs=-1,\n objective=None,\n subsample=0.7, \n subsample_for_bin=240000, \n subsample_freq=0)\n\nlgbm_model_3 = lgb.LGBMClassifier(boosting_type='gbdt', \n colsample_bytree=1.0,\n importance_type='split', \n learning_rate=0.20,\n max_depth=40,\n n_estimators=1450, \n n_jobs=-1,\n objective=None,\n subsample=0.7, \n subsample_for_bin=160000, \n subsample_freq=0)",
"_____no_output_____"
],
[
"import sklearn as sk\nimport sklearn.neural_network\nneuronal_1 = sk.neural_network.MLPClassifier(solver='adam', \n activation = 'relu',\n learning_rate_init=0.001,\n learning_rate = 'adaptive',\n verbose=True,\n batch_size = 'auto')",
"_____no_output_____"
],
[
"gb_model_1 = GradientBoostingClassifier(n_estimators = 305,\n max_depth = 9,\n min_samples_split = 2,\n min_samples_leaf = 3,\n subsample=0.6,\n verbose=True,\n learning_rate=0.15)",
"_____no_output_____"
],
[
"vc_model = VotingClassifier(estimators = [('xgb1', xgb_model_1),\n ('xgb2', xgb_model_2),\n ('rfm1', rf_model_1),\n ('lgbm1', lgbm_model_1),\n ('lgbm2', lgbm_model_2),\n ('gb_model_1', gb_model_1)],\n weights = [1.0, 0.95, 0.85, 1.0, 0.9, 0.7, 0.9],\n voting = 'soft',\n verbose = True)",
"_____no_output_____"
],
[
"vc_model.fit(X_train, y_train)",
"/home/joaquinfontela/.local/lib/python3.8/site-packages/xgboost/sklearn.py:1146: UserWarning: The use of label encoder in XGBClassifier is deprecated and will be removed in a future release. To remove this warning, do the following: 1) Pass option use_label_encoder=False when constructing XGBClassifier object; and 2) Encode your labels (y) as integers starting with 0, i.e. 0, 1, 2, ..., [num_class - 1].\n warnings.warn(label_encoder_deprecation_msg, UserWarning)\n"
],
[
"y_preds = vc_model.predict(X_test)\nf1_score(y_test, y_preds, average='micro')",
"[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.\n[Parallel(n_jobs=1)]: Done 150 out of 150 | elapsed: 2.1s finished\n"
],
[
"test_values = pd.read_csv('../../csv/test_values.csv', index_col = \"building_id\")\ntest_values",
"_____no_output_____"
],
[
"test_values_subset = test_values\ntest_values_subset[\"geo_level_1_id\"] = test_values_subset[\"geo_level_1_id\"].astype(\"category\")\ntest_values_subset",
"_____no_output_____"
],
[
"def encode_and_bind(original_dataframe, feature_to_encode):\n dummies = pd.get_dummies(original_dataframe[[feature_to_encode]])\n res = pd.concat([original_dataframe, dummies], axis=1)\n res = res.drop([feature_to_encode], axis=1)\n return(res) \n\nfeatures_to_encode = [\"geo_level_1_id\", \"land_surface_condition\", \"foundation_type\", \"roof_type\",\\\n \"position\", \"ground_floor_type\", \"other_floor_type\",\\\n \"plan_configuration\", \"legal_ownership_status\"]\nfor feature in features_to_encode:\n test_values_subset = encode_and_bind(test_values_subset, feature)\ntest_values_subset",
"_____no_output_____"
],
[
"test_values_subset.shape",
"_____no_output_____"
],
[
"# Genero las predicciones para los test.\npreds = vc_model.predict(test_values_subset)",
"[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.\n[Parallel(n_jobs=1)]: Done 150 out of 150 | elapsed: 3.7s finished\n[Parallel(n_jobs=8)]: Using backend ThreadingBackend with 8 concurrent workers.\n[Parallel(n_jobs=8)]: Done 34 tasks | elapsed: 0.3s\n[Parallel(n_jobs=8)]: Done 184 tasks | elapsed: 1.4s\n[Parallel(n_jobs=8)]: Done 250 out of 250 | elapsed: 1.8s finished\n"
],
[
"submission_format = pd.read_csv('../../csv/submission_format.csv', index_col = \"building_id\")",
"_____no_output_____"
],
[
"my_submission = pd.DataFrame(data=preds,\n columns=submission_format.columns,\n index=submission_format.index)",
"_____no_output_____"
],
[
"my_submission.head()",
"_____no_output_____"
],
[
"my_submission.to_csv('../../csv/predictions/jf/vote/jf-model-3-submission.csv')",
"_____no_output_____"
],
[
"!head ../../csv/predictions/jf/vote/jf-model-3-submission.csv",
"building_id,damage_grade\r\n300051,3\r\n99355,2\r\n890251,2\r\n745817,1\r\n421793,3\r\n871976,2\r\n691228,1\r\n896100,3\r\n343471,2\r\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d000c817d7ae0508e04ac21d6d2f4e30cbc917d4 | 146,114 | ipynb | Jupyter Notebook | Python_Stock/Time_Series_Forecasting/Stock_Forecasting_Prophet_Uncertainty_Trend.ipynb | LastAncientOne/Stock_Analysis_For_Quant | b21d01d0fdd0098e454147942d9b07979ab315ad | [
"MIT"
] | 962 | 2019-07-17T09:57:41.000Z | 2022-03-29T01:55:20.000Z | Python_Stock/Time_Series_Forecasting/Stock_Forecasting_Prophet_Uncertainty_Trend.ipynb | j0el/Stock_Analysis_For_Quant | 8088fb0f6a1b1edeead6ae152fa4275e3d6dd746 | [
"MIT"
] | 5 | 2020-04-29T16:54:30.000Z | 2022-02-10T02:57:30.000Z | Python_Stock/Time_Series_Forecasting/Stock_Forecasting_Prophet_Uncertainty_Trend.ipynb | j0el/Stock_Analysis_For_Quant | 8088fb0f6a1b1edeead6ae152fa4275e3d6dd746 | [
"MIT"
] | 286 | 2019-08-04T10:37:58.000Z | 2022-03-28T06:31:56.000Z | 363.467662 | 81,973 | 0.905321 | [
[
[
"# Stock Forecasting using Prophet (Uncertainty in the trend)",
"_____no_output_____"
],
[
"https://facebook.github.io/prophet/",
"_____no_output_____"
]
],
[
[
"# Libraries\n\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom prophet import Prophet\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\nimport yfinance as yf\nyf.pdr_override()",
"_____no_output_____"
],
[
"stock = 'AMD' # input\nstart = '2017-01-01' # input \nend = '2021-11-08' # input\ndf = yf.download(stock, start, end)",
"[*********************100%***********************] 1 of 1 completed\n"
],
[
"plt.figure(figsize=(16,8))\nplt.plot(df['Adj Close'])\nplt.title('Stock Price')\nplt.ylabel('Price')\nplt.show()",
"_____no_output_____"
],
[
"df = df.reset_index()\ndf = df.rename(columns={'Date': 'ds', 'Close': 'y'})\ndf",
"_____no_output_____"
],
[
"df = df[['ds', 'y']]\ndf",
"_____no_output_____"
],
[
"m = Prophet(daily_seasonality=True)\nm.fit(df)",
"_____no_output_____"
],
[
"future = m.make_future_dataframe(periods=365)\nfuture.tail()",
"_____no_output_____"
],
[
"m = Prophet(mcmc_samples=300)\nforecast = m.fit(df).predict(future)\n",
"INFO:prophet:Disabling daily seasonality. Run prophet with daily_seasonality=True to override this.\nWARNING:pystan:Maximum (flat) parameter count (1000) exceeded: skipping diagnostic tests for n_eff and Rhat.\nTo run all diagnostics call pystan.check_hmc_diagnostics(fit)\nWARNING:pystan:597 of 600 iterations saturated the maximum tree depth of 10 (99.5 %)\nWARNING:pystan:Run again with max_treedepth larger than 10 to avoid saturation\n"
],
[
"fig = m.plot_components(forecast)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d000dbbbe2638cb9586ae564f2c03042b309708f | 7,478 | ipynb | Jupyter Notebook | delfin/Example - Delfin.ipynb | Open-Dataplatform/examples | 197e83308bef83b09a32fea898f16c8cd9c84acb | [
"MIT"
] | null | null | null | delfin/Example - Delfin.ipynb | Open-Dataplatform/examples | 197e83308bef83b09a32fea898f16c8cd9c84acb | [
"MIT"
] | null | null | null | delfin/Example - Delfin.ipynb | Open-Dataplatform/examples | 197e83308bef83b09a32fea898f16c8cd9c84acb | [
"MIT"
] | null | null | null | 26.424028 | 141 | 0.514175 | [
[
[
"# Delfin",
"_____no_output_____"
],
[
"### Installation\nRun the following cell to install osiris-sdk.",
"_____no_output_____"
]
],
[
[
"!pip install osiris-sdk --upgrade",
"_____no_output_____"
]
],
[
[
"### Access to dataset\nThere are two ways to get access to a dataset\n1. Service Principle\n2. Access Token\n\n\n#### Config file with Service Principle\nIf done with **Service Principle** it is adviced to add the following file with **tenant_id**, **client_id**, and **client_secret**:\n\nThe structure of **conf.ini**:\n```\n[Authorization]\ntenant_id = <tenant_id>\nclient_id = <client_id>\nclient_secret = <client_secret>\n\n[Egress]\nurl = <egress-url>\n```\n\n#### Config file if using Access Token\nIf done with **Access Token** then assign it to a variable (see example below).\n\nThe structure of **conf.ini**:\n```\n[Egress]\nurl = <egress-url>\n```\n\nThe egress-url can be [found here](https://github.com/Open-Dataplatform/examples/blob/main/README.md).",
"_____no_output_____"
],
[
"### Imports\nExecute the following cell to import the necessary libraries",
"_____no_output_____"
]
],
[
[
"from osiris.apis.egress import Egress\nfrom osiris.core.azure_client_authorization import ClientAuthorization\nfrom osiris.core.enums import Horizon\nfrom configparser import ConfigParser",
"_____no_output_____"
]
],
[
[
"### Initialize the Egress class with Service Principle",
"_____no_output_____"
]
],
[
[
"config = ConfigParser()\nconfig.read('conf.ini')\n\nclient_auth = ClientAuthorization(tenant_id=config['Authorization']['tenant_id'],\n client_id=config['Authorization']['client_id'],\n client_secret=config['Authorization']['client_secret'])\n\negress = Egress(client_auth=client_auth,\n egress_url=config['Egress']['url'])",
"_____no_output_____"
]
],
[
[
"### Intialize the Egress class with Access Token",
"_____no_output_____"
]
],
[
[
"config = ConfigParser()\nconfig.read('conf.ini')\n\naccess_token = 'REPLACE WITH ACCESS TOKEN HERE'\n\nclient_auth = ClientAuthorization(access_token=access_token)\n\negress = Egress(client_auth=client_auth,\n egress_url=config['Egress']['url'])",
"_____no_output_____"
]
],
[
[
"### Delfin Daily\nThe data retrived will be **from_date <= data < to_date**.\n\nThe **from_date** and **to_date** syntax is [described here](https://github.com/Open-Dataplatform/examples/blob/main/README.md).",
"_____no_output_____"
]
],
[
[
"json_content = egress.download_delfin_file(horizon=Horizon.MINUTELY, \n from_date=\"2021-07-15T20:00\", \n to_date=\"2021-07-16T00:00\")",
"_____no_output_____"
],
[
"json_content = egress.download_delfin_file(horizon=Horizon.DAILY, \n from_date=\"2020-01\", \n to_date=\"2020-02\")\n\n# We only show the first entry here\njson_content[0]",
"_____no_output_____"
]
],
[
[
"### Delfin Hourly\nThe **from_date** and **to_date** syntax is [described here](https://github.com/Open-Dataplatform/examples/blob/main/README.md).",
"_____no_output_____"
]
],
[
[
"json_content = egress.download_delfin_file(horizon=Horizon.HOURLY, \n from_date=\"2020-01-01T00\", \n to_date=\"2020-01-01T06\")\n\n# We only show the first entry here\njson_content[0]",
"_____no_output_____"
]
],
[
[
"### Delfin Minutely\nThe **from_date** and **to_date** syntax is [described here](https://github.com/Open-Dataplatform/examples/blob/main/README.md).",
"_____no_output_____"
]
],
[
[
"json_content = egress.download_delfin_file(horizon=Horizon.MINUTELY, \n from_date=\"2021-07-15T00:00\", \n to_date=\"2021-07-15T00:59\")\n\n# We only show the first entry here\njson_content[0]",
"_____no_output_____"
]
],
[
[
"### Delfin Daily with Indices\nThe **from_date** and **to_date** syntax is [described here](https://github.com/Open-Dataplatform/examples/blob/main/README.md).",
"_____no_output_____"
]
],
[
[
"json_content = egress.download_delfin_file(horizon=Horizon.DAILY, \n from_date=\"2020-01-15T03:00\", \n to_date=\"2020-01-16T03:01\",\n table_indices=[1, 2])\n\n# We only show the first entry here\njson_content[0]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d000df8fd6872f4ebeceecda2014c8ed69838b8d | 49,315 | ipynb | Jupyter Notebook | 09_Time_Series/Apple_Stock/Exercises-with-solutions-code.ipynb | nat-bautista/tts-pandas-exercise | dd288b691e1789801b76675fed581c854adfaa26 | [
"BSD-3-Clause"
] | null | null | null | 09_Time_Series/Apple_Stock/Exercises-with-solutions-code.ipynb | nat-bautista/tts-pandas-exercise | dd288b691e1789801b76675fed581c854adfaa26 | [
"BSD-3-Clause"
] | null | null | null | 09_Time_Series/Apple_Stock/Exercises-with-solutions-code.ipynb | nat-bautista/tts-pandas-exercise | dd288b691e1789801b76675fed581c854adfaa26 | [
"BSD-3-Clause"
] | null | null | null | 70.956835 | 31,322 | 0.743303 | [
[
[
"# Apple Stock",
"_____no_output_____"
],
[
"### Introduction:\n\nWe are going to use Apple's stock price.\n\n\n### Step 1. Import the necessary libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\n# visualization\nimport matplotlib.pyplot as plt\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Step 2. Import the dataset from this [address](https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/09_Time_Series/Apple_Stock/appl_1980_2014.csv)",
"_____no_output_____"
],
[
"### Step 3. Assign it to a variable apple",
"_____no_output_____"
]
],
[
[
"url = 'https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/09_Time_Series/Apple_Stock/appl_1980_2014.csv'\napple = pd.read_csv(url)\n\napple.head()",
"_____no_output_____"
]
],
[
[
"### Step 4. Check out the type of the columns",
"_____no_output_____"
]
],
[
[
"apple.dtypes",
"_____no_output_____"
]
],
[
[
"### Step 5. Transform the Date column as a datetime type",
"_____no_output_____"
]
],
[
[
"apple.Date = pd.to_datetime(apple.Date)\n\napple['Date'].head()",
"_____no_output_____"
]
],
[
[
"### Step 6. Set the date as the index",
"_____no_output_____"
]
],
[
[
"apple = apple.set_index('Date')\n\napple.head()",
"_____no_output_____"
]
],
[
[
"### Step 7. Is there any duplicate dates?",
"_____no_output_____"
]
],
[
[
"# NO! All are unique\napple.index.is_unique",
"_____no_output_____"
]
],
[
[
"### Step 8. Ops...it seems the index is from the most recent date. Make the first entry the oldest date.",
"_____no_output_____"
]
],
[
[
"apple.sort_index(ascending = True).head()",
"_____no_output_____"
]
],
[
[
"### Step 9. Get the last business day of each month",
"_____no_output_____"
]
],
[
[
"apple_month = apple.resample('BM').mean()\n\napple_month.head()",
"_____no_output_____"
]
],
[
[
"### Step 10. What is the difference in days between the first day and the oldest",
"_____no_output_____"
]
],
[
[
"(apple.index.max() - apple.index.min()).days",
"_____no_output_____"
]
],
[
[
"### Step 11. How many months in the data we have?",
"_____no_output_____"
]
],
[
[
"apple_months = apple.resample('BM').mean()\n\nlen(apple_months.index)",
"_____no_output_____"
]
],
[
[
"### Step 12. Plot the 'Adj Close' value. Set the size of the figure to 13.5 x 9 inches",
"_____no_output_____"
]
],
[
[
"# makes the plot and assign it to a variable\nappl_open = apple['Adj Close'].plot(title = \"Apple Stock\")\n\n# changes the size of the graph\nfig = appl_open.get_figure()\nfig.set_size_inches(13.5, 9)",
"_____no_output_____"
]
],
[
[
"### BONUS: Create your own question and answer it.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d000ef77a4fe5d63f756fa5540228432b5512788 | 13,227 | ipynb | Jupyter Notebook | Colab RDP/Colab RDP.ipynb | Apon77/Colab-Hacks | 3493aa482b7420b8f7c2d236308dd3568254860c | [
"MIT"
] | null | null | null | Colab RDP/Colab RDP.ipynb | Apon77/Colab-Hacks | 3493aa482b7420b8f7c2d236308dd3568254860c | [
"MIT"
] | null | null | null | Colab RDP/Colab RDP.ipynb | Apon77/Colab-Hacks | 3493aa482b7420b8f7c2d236308dd3568254860c | [
"MIT"
] | 2 | 2021-02-24T20:42:46.000Z | 2021-04-22T01:14:30.000Z | 37.791429 | 644 | 0.482422 | [
[
[
"<a href=\"https://colab.research.google.com/github/PradyumnaKrishna/Colab-Hacks/blob/RDP-v2/Colab%20RDP/Colab%20RDP.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# **Colab RDP** : Remote Desktop to Colab Instance\n\nUsed Google Remote Desktop & Ngrok Tunnel\n\n> **Warning : Not for Cryptocurrency Mining<br></br>** \n>**Why are hardware resources such as T4 GPUs not available to me?** The best available hardware is prioritized for users who use Colaboratory interactively rather than for long-running computations. Users who use Colaboratory for long-running computations may be temporarily restricted in the type of hardware made available to them, and/or the duration that the hardware can be used for. We encourage users with high computational needs to use Colaboratory’s UI with a local runtime. Please note that using Colaboratory for cryptocurrency mining is disallowed entirely, and may result in being banned from using Colab altogether.\n\nGoogle Colab can give you Instance with 12GB of RAM and GPU for 12 hours (Max.) for Free users. Anyone can use it to perform Heavy Tasks.\n\nTo use other similiar Notebooks use my Repository **[Colab Hacks](https://github.com/PradyumnaKrishna/Colab-Hacks)**",
"_____no_output_____"
]
],
[
[
"#@title **Create User**\n#@markdown Enter Username and Password\n\nusername = \"user\" #@param {type:\"string\"}\npassword = \"root\" #@param {type:\"string\"}\n\nprint(\"Creating User and Setting it up\")\n\n# Creation of user\n! sudo useradd -m $username &> /dev/null\n\n# Add user to sudo group\n! sudo adduser $username sudo &> /dev/null\n \n# Set password of user to 'root'\n! echo '$username:$password' | sudo chpasswd\n\n# Change default shell from sh to bash\n! sed -i 's/\\/bin\\/sh/\\/bin\\/bash/g' /etc/passwd\n\nprint(\"User Created and Configured\")",
"_____no_output_____"
],
[
"#@title **RDP**\n#@markdown It takes 4-5 minutes for installation\n\n#@markdown Visit http://remotedesktop.google.com/headless and Copy the command after authentication\n\nCRP = \"\" #@param {type:\"string\"}\n\ndef CRD():\n with open('install.sh', 'w') as script:\n script.write(\"\"\"#! /bin/bash\n\nb='\\033[1m'\nr='\\E[31m'\ng='\\E[32m'\nc='\\E[36m'\nendc='\\E[0m'\nenda='\\033[0m'\n\nprintf \"\\n\\n$c$b Loading Installer $endc$enda\" >&2\nif sudo apt-get update &> /dev/null\nthen\n printf \"\\r$g$b Installer Loaded $endc$enda\\n\" >&2\nelse\n printf \"\\r$r$b Error Occured $endc$enda\\n\" >&2\n exit\nfi\n\nprintf \"\\n$g$b Installing Chrome Remote Desktop $endc$enda\" >&2\n{\n wget https://dl.google.com/linux/direct/chrome-remote-desktop_current_amd64.deb\n sudo dpkg --install chrome-remote-desktop_current_amd64.deb\n sudo apt install --assume-yes --fix-broken\n} &> /dev/null &&\nprintf \"\\r$c$b Chrome Remote Desktop Installed $endc$enda\\n\" >&2 ||\n{ printf \"\\r$r$b Error Occured $endc$enda\\n\" >&2; exit; }\nsleep 3\n\nprintf \"$g$b Installing Desktop Environment $endc$enda\" >&2\n{\n sudo DEBIAN_FRONTEND=noninteractive \\\n apt install --assume-yes xfce4 desktop-base\n sudo bash -c 'echo \"exec /etc/X11/Xsession /usr/bin/xfce4-session\" > /etc/chrome-remote-desktop-session' \n sudo apt install --assume-yes xscreensaver\n sudo systemctl disable lightdm.service\n} &> /dev/null &&\nprintf \"\\r$c$b Desktop Environment Installed $endc$enda\\n\" >&2 ||\n{ printf \"\\r$r$b Error Occured $endc$enda\\n\" >&2; exit; }\nsleep 3\n\nprintf \"$g$b Installing Google Chrome $endc$enda\" >&2\n{\n wget https://dl.google.com/linux/direct/google-chrome-stable_current_amd64.deb\n sudo dpkg --install google-chrome-stable_current_amd64.deb\n sudo apt install --assume-yes --fix-broken\n} &> /dev/null &&\nprintf \"\\r$c$b Google Chrome Installed $endc$enda\\n\" >&2 ||\nprintf \"\\r$r$b Error Occured $endc$enda\\n\" >&2\nsleep 3\n\nprintf \"$g$b Installing other Tools $endc$enda\" >&2\nif sudo apt install nautilus nano -y &> /dev/null\nthen\n printf \"\\r$c$b Other Tools Installed $endc$enda\\n\" >&2\nelse\n printf \"\\r$r$b Error Occured $endc$enda\\n\" >&2\nfi\nsleep 3\n\nprintf \"\\n$g$b Installation Completed $endc$enda\\n\\n\" >&2\"\"\")\n\n ! chmod +x install.sh\n ! ./install.sh\n\n # Adding user to CRP group\n ! sudo adduser $username chrome-remote-desktop &> /dev/null\n\n # Finishing Work\n ! su - $username -c \"\"\"$CRP\"\"\"\n\n print(\"Finished Succesfully\")\n\ntry:\n if username:\n if CRP == \"\" :\n print(\"Please enter authcode from the given link\")\n else:\n CRD()\nexcept NameError:\n print(\"username variable not found\")\n print(\"Create a User First\")",
"_____no_output_____"
],
[
"#@title **Google Drive Mount**\n#@markdown Google Drive used as Persistance HDD for files.<br>\n#@markdown Mounted at `user` Home directory inside drive folder\n#@markdown (If `username` variable not defined then use root as default).\n\ndef MountGDrive():\n from google.colab import drive\n\n ! runuser -l $user -c \"yes | python3 -m pip install --user google-colab\" > /dev/null 2>&1\n\n mount = \"\"\"from os import environ as env\nfrom google.colab import drive\n\nenv['CLOUDSDK_CONFIG'] = '/content/.config'\ndrive.mount('{}')\"\"\".format(mountpoint)\n\n with open('/content/mount.py', 'w') as script:\n script.write(mount)\n\n ! runuser -l $user -c \"python3 /content/mount.py\"\n\ntry:\n if username:\n mountpoint = \"/home/\"+username+\"/drive\"\n user = username\nexcept NameError:\n print(\"username variable not found, mounting at `/content/drive' using `root'\")\n mountpoint = '/content/drive'\n user = 'root'\n\nMountGDrive()",
"_____no_output_____"
],
[
"#@title **SSH** (Using NGROK)\n\n! pip install colab_ssh --upgrade &> /dev/null\nfrom colab_ssh import launch_ssh, init_git\nfrom IPython.display import clear_output\n\n#@markdown Copy authtoken from https://dashboard.ngrok.com/auth\nngrokToken = \"\" #@param {type:'string'}\n\ndef runNGROK():\n launch_ssh(ngrokToken, password)\n clear_output()\n\n print(\"ssh\", username, end='@')\n ! curl -s http://localhost:4040/api/tunnels | python3 -c \\\n \"import sys, json; print(json.load(sys.stdin)['tunnels'][0]['public_url'][6:].replace(':', ' -p '))\"\n\ntry:\n if username:\n pass\n elif password:\n pass\nexcept NameError:\n print(\"No user found using username and password as 'root'\")\n username='root'\n password='root'\n\nif ngrokToken == \"\":\n print(\"No ngrokToken Found, Please enter it\")\nelse:\n runNGROK()",
"_____no_output_____"
],
[
"#@title Package Installer { vertical-output: true }\nrun = False #@param {type:\"boolean\"}\n#@markdown *Package management actions (gasp)*\naction = \"Install\" #@param [\"Install\", \"Check Installed\", \"Remove\"] {allow-input: true}\n\npackage = \"wget\" #@param {type:\"string\"}\nsystem = \"apt\" #@param [\"apt\", \"\"]\n\ndef install(package=package, system=system):\n if system == \"apt\":\n !apt --fix-broken install > /dev/null 2>&1\n !killall apt > /dev/null 2>&1\n !rm /var/lib/dpkg/lock-frontend\n !dpkg --configure -a > /dev/null 2>&1\n\n !apt-get install -o Dpkg::Options::=\"--force-confold\" --no-install-recommends -y $package\n \n !dpkg --configure -a > /dev/null 2>&1 \n !apt update > /dev/null 2>&1\n\n !apt install $package > /dev/null 2>&1\n\ndef check_installed(package=package, system=system):\n if system == \"apt\":\n !apt list --installed | grep $package\n\ndef remove(package=package, system=system):\n if system == \"apt\":\n !apt remove $package\n\nif run:\n if action == \"Install\":\n install()\n if action == \"Check Installed\":\n check_installed()\n if action == \"Remove\":\n remove()",
"_____no_output_____"
],
[
"#@title **Colab Shutdown**\n\n#@markdown To Kill NGROK Tunnel\nNGROK = False #@param {type:'boolean'}\n\n#@markdown To Unmount GDrive\nGDrive = False #@param {type:'boolean'}\n\n#@markdown To Sleep Colab\nSleep = False #@param {type:'boolean'}\n\nif NGROK:\n ! killall ngrok\n\nif GDrive:\n with open('/content/unmount.py', 'w') as unmount:\n unmount.write(\"\"\"from google.colab import drive\ndrive.flush_and_unmount()\"\"\")\n \n try:\n if user:\n ! runuser $user -c 'python3 /content/unmount.py'\n except NameError:\n print(\"Google Drive not Mounted\")\n\nif Sleep:\n ! sleep 43200",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d000f1ce0f008b8f64f705810da78b9e62f26064 | 63,150 | ipynb | Jupyter Notebook | scanpy_cellphonedb.ipynb | stefanpeidli/cellphonedb | c638935d7fc36e0c3156a1a8c26d2e0108b2bf0e | [
"MIT"
] | null | null | null | scanpy_cellphonedb.ipynb | stefanpeidli/cellphonedb | c638935d7fc36e0c3156a1a8c26d2e0108b2bf0e | [
"MIT"
] | null | null | null | scanpy_cellphonedb.ipynb | stefanpeidli/cellphonedb | c638935d7fc36e0c3156a1a8c26d2e0108b2bf0e | [
"MIT"
] | 1 | 2021-02-03T16:25:06.000Z | 2021-02-03T16:25:06.000Z | 45.398994 | 212 | 0.357846 | [
[
[
"from IPython.core.display import display, HTML\ndisplay(HTML(\"<style>.container { width:90% !important; }</style>\"))\n%matplotlib inline\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as pl\nimport scanpy as sc\n\nimport cellphonedb as cphdb\n\n# Original API works for python as well, it's just not really nice\nimport sys \nsys.path.insert(0, './cellphonedb/src/api_endpoints/terminal_api/method_terminal_api_endpoints/')\nfrom method_terminal_commands import statistical_analysis",
"_____no_output_____"
]
],
[
[
"# Dev",
"_____no_output_____"
],
[
"## Original method",
"_____no_output_____"
]
],
[
[
"# you need to download these from cellphonedb website / github and replace the path accordingly\ndat = 'C:/Users/Stefan/Downloads/cellphonedb_example_data/example_data/'\nmetafile = dat+'test_meta.txt'\ncountfile = dat+'test_counts.txt'\n\nstatistical_analysis(meta_filename=metafile, counts_filename=countfile)",
"[ ][APP][04/11/20-17:12:40][WARNING] Latest local available version is `v2.0.0`, using it\n[ ][APP][04/11/20-17:12:40][WARNING] User selected downloaded database `v2.0.0` is available, using it\n[ ][CORE][04/11/20-17:12:40][INFO] Initializing SqlAlchemy CellPhoneDB Core\n[ ][CORE][04/11/20-17:12:40][INFO] Using custom database at C:\\Users\\Stefan\\.cpdb\\releases\\v2.0.0\\cellphone.db\n[ ][APP][04/11/20-17:12:40][INFO] Launching Method cpdb_statistical_analysis_local_method_launcher\n[ ][APP][04/11/20-17:12:40][INFO] Launching Method _set_paths\n[ ][APP][04/11/20-17:12:40][WARNING] Output directory (C:\\Users\\Stefan\\Documents\\Github_Clones\\cellphonedb/out) exist and is not empty. Result can overwrite old results\n[ ][APP][04/11/20-17:12:40][INFO] Launching Method _load_meta_counts\n[ ][CORE][04/11/20-17:12:40][INFO] Launching Method cpdb_statistical_analysis_launcher\n[ ][CORE][04/11/20-17:12:40][INFO] Launching Method _counts_validations\n[ ][CORE][04/11/20-17:12:40][INFO] [Cluster Statistical Analysis Simple] Threshold:0.1 Iterations:1000 Debug-seed:-1 Threads:4 Precision:3\n[ ][CORE][04/11/20-17:12:40][INFO] Running Simple Prefilters\n[ ][CORE][04/11/20-17:12:40][INFO] Running Real Simple Analysis\n[ ][CORE][04/11/20-17:12:40][INFO] Running Statistical Analysis\n[ ][CORE][04/11/20-17:13:27][INFO] Building Pvalues result\n[ ][CORE][04/11/20-17:13:29][INFO] Building Simple results\n[ ][CORE][04/11/20-17:13:29][INFO] [Cluster Statistical Analysis Complex] Threshold:0.1 Iterations:1000 Debug-seed:-1 Threads:4 Precision:3\n[ ][CORE][04/11/20-17:13:29][INFO] Running Complex Prefilters\n[ ][CORE][04/11/20-17:13:31][INFO] Running Real Complex Analysis\n[ ][CORE][04/11/20-17:13:32][INFO] Running Statistical Analysis\n[ ][CORE][04/11/20-17:14:38][INFO] Building Pvalues result\n[ ][CORE][04/11/20-17:14:40][INFO] Building Complex results\n"
],
[
"pd.read_csv('./out/pvalues.csv')",
"_____no_output_____"
]
],
[
[
"## scanpy API test official cellphonedb example data",
"_____no_output_____"
]
],
[
[
"# you need to download these from cellphonedb website / github and replace the path accordingly\ndat = 'C:/Users/Stefan/Downloads/cellphonedb_example_data/example_data/'\nmetafile = dat+'test_meta.txt'\ncountfile = dat+'test_counts.txt'",
"_____no_output_____"
],
[
"bdata=sc.AnnData(pd.read_csv(countfile, sep='\\t',index_col=0).values.T, obs=pd.read_csv(metafile, sep='\\t',index_col=0), var=pd.DataFrame(index=pd.read_csv(countfile, sep='\\t',index_col=0).index.values))",
"_____no_output_____"
],
[
"outs=cphdb.statistical_analysis_scanpy(bdata, bdata.var_names, bdata.obs_names, 'cell_type')",
"[ ][APP][04/11/20-17:14:43][WARNING] Latest local available version is `v2.0.0`, using it\n[ ][APP][04/11/20-17:14:43][WARNING] User selected downloaded database `v2.0.0` is available, using it\n[ ][CORE][04/11/20-17:14:43][INFO] Initializing SqlAlchemy CellPhoneDB Core\n[ ][CORE][04/11/20-17:14:43][INFO] Using custom database at C:\\Users\\Stefan\\.cpdb\\releases\\v2.0.0\\cellphone.db\n[ ][APP][04/11/20-17:14:43][INFO] Launching Method cpdb_statistical_analysis_local_method_launcher_scanpy\n[ ][APP][04/11/20-17:14:43][INFO] Launching Method _set_paths\n[ ][APP][04/11/20-17:14:43][WARNING] Output directory (C:\\Users\\Stefan\\Documents\\Github_Clones\\cellphonedb/out) exist and is not empty. Result can overwrite old results\n[ ][CORE][04/11/20-17:14:43][INFO] Launching Method cpdb_statistical_analysis_launcher\n[ ][CORE][04/11/20-17:14:43][INFO] Launching Method _counts_validations\n[ ][CORE][04/11/20-17:14:43][INFO] [Cluster Statistical Analysis Simple] Threshold:0.1 Iterations:1000 Debug-seed:-1 Threads:4 Precision:3\n[ ][CORE][04/11/20-17:14:43][INFO] Running Simple Prefilters\n[ ][CORE][04/11/20-17:14:43][INFO] Running Real Simple Analysis\n[ ][CORE][04/11/20-17:14:43][INFO] Running Statistical Analysis\n[ ][CORE][04/11/20-17:15:32][INFO] Building Pvalues result\n[ ][CORE][04/11/20-17:15:35][INFO] Building Simple results\n[ ][CORE][04/11/20-17:15:35][INFO] [Cluster Statistical Analysis Complex] Threshold:0.1 Iterations:1000 Debug-seed:-1 Threads:4 Precision:3\n[ ][CORE][04/11/20-17:15:35][INFO] Running Complex Prefilters\n[ ][CORE][04/11/20-17:15:37][INFO] Running Real Complex Analysis\n[ ][CORE][04/11/20-17:15:38][INFO] Running Statistical Analysis\n[ ][CORE][04/11/20-17:16:38][INFO] Building Pvalues result\n[ ][CORE][04/11/20-17:16:39][INFO] Building Complex results\n"
],
[
"outs['pvalues']",
"_____no_output_____"
],
[
"# the output is also saved to\nbdata.uns['cellphonedb_output']",
"_____no_output_____"
],
[
"bdata",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d000fa08beeccd71f734c998515820b69c9a44b2 | 35,623 | ipynb | Jupyter Notebook | isis/notebooks/crop_eis.ipynb | gknorman/ISIS3 | 4800a8047626a864e163cc74055ba60008c105f7 | [
"CC0-1.0"
] | 134 | 2018-01-18T00:16:24.000Z | 2022-03-24T03:53:33.000Z | isis/notebooks/crop_eis.ipynb | gknorman/ISIS3 | 4800a8047626a864e163cc74055ba60008c105f7 | [
"CC0-1.0"
] | 3,825 | 2017-12-11T21:27:34.000Z | 2022-03-31T21:45:20.000Z | isis/notebooks/crop_eis.ipynb | jessemapel/ISIS3 | bd43b627378c4009c6aaae8537ba472dbefb2152 | [
"CC0-1.0"
] | 164 | 2017-11-30T21:15:44.000Z | 2022-03-23T10:22:29.000Z | 30.291667 | 5,000 | 0.4318 | [
[
[
"from xml.dom import expatbuilder\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport struct\nimport os\n",
"_____no_output_____"
],
[
"# should be in the same directory as corresponding xml and csv\neis_filename = '/example/path/to/eis_image_file.dat'",
"_____no_output_____"
],
[
"image_fn, image_ext = os.path.splitext(eis_filename)\neis_xml_filename = image_fn + \".xml\"",
"_____no_output_____"
]
],
[
[
"# crop xml",
"_____no_output_____"
],
[
"manually change the line and sample values in the xml to match (n_lines, n_samples)",
"_____no_output_____"
]
],
[
[
"eis_xml = expatbuilder.parse(eis_xml_filename, False)",
"_____no_output_____"
],
[
"eis_dom = eis_xml.getElementsByTagName(\"File_Area_Observational\").item(0)\n\ndom_lines = eis_dom.getElementsByTagName(\"Axis_Array\").item(0)\ndom_samples = eis_dom.getElementsByTagName(\"Axis_Array\").item(1)\n\ndom_lines = dom_lines.getElementsByTagName(\"elements\")[0]\ndom_samples = dom_samples.getElementsByTagName(\"elements\")[0]\n\ntotal_lines = int( dom_lines.childNodes[0].data )\ntotal_samples = int( dom_samples.childNodes[0].data )\n\ntotal_lines, total_samples",
"_____no_output_____"
]
],
[
[
"# crop image",
"_____no_output_____"
]
],
[
[
"\ndn_size_bytes = 4 # number of bytes per DN\n\nn_lines = 60 # how many to crop to\nn_samples = 3\n\nstart_line = 1200 # point to start crop from\nstart_sample = 1200\n\nimage_offset = (start_line*total_samples + start_sample) * dn_size_bytes\n\nline_length = n_samples * dn_size_bytes\n\nbuffer_size = n_lines * total_samples * dn_size_bytes\n\n\nwith open(eis_filename, 'rb') as f:\n f.seek(image_offset) \n b_image_data = f.read()",
"_____no_output_____"
],
[
"b_image_data = np.frombuffer(b_image_data[:buffer_size], dtype=np.uint8)\nb_image_data.shape",
"_____no_output_____"
],
[
"b_image_data = np.reshape(b_image_data, (n_lines, total_samples, dn_size_bytes) )\nb_image_data.shape",
"_____no_output_____"
],
[
"b_image_data = b_image_data[:,:n_samples,:]\nb_image_data.shape",
"_____no_output_____"
],
[
"image_data = []\nfor i in range(n_lines):\n image_sample = []\n for j in range(n_samples):\n dn_bytes = bytearray(b_image_data[i,j,:])\n dn = struct.unpack( \"<f\", dn_bytes )\n image_sample.append(dn)\n image_data.append(image_sample)\nimage_data = np.array(image_data)\nimage_data.shape",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,10))\nplt.imshow(image_data, vmin=0, vmax=1)",
"_____no_output_____"
],
[
"crop = \"_cropped\"\nimage_fn, image_ext = os.path.splitext(eis_filename)\nmini_image_fn = image_fn + crop + image_ext\nmini_image_bn = os.path.basename(mini_image_fn)",
"_____no_output_____"
],
[
"if os.path.exists(mini_image_fn):\n os.remove(mini_image_fn)\n\nwith open(mini_image_fn, 'ab+') as f:\n b_reduced_image_data = image_data.tobytes()\n f.write(b_reduced_image_data)",
"_____no_output_____"
]
],
[
[
"# crop times csv table",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\n# assumes csv file has the same filename with _times appended\neis_csv_fn = image_fn + \"_times.csv\"\ndf1 = pd.read_csv(eis_csv_fn)\ndf1",
"_____no_output_____"
],
[
"x = np.array(df1)\ny = x[:n_lines, :]\ndf = pd.DataFrame(y)\ndf",
"_____no_output_____"
],
[
"crop = \"_cropped\"\ncsv_fn, csv_ext = os.path.splitext(eis_csv_fn)\ncrop_csv_fn = csv_fn + crop + csv_ext\ncrop_csv_bn = os.path.basename(crop_csv_fn)\ncrop_csv_bn",
"_____no_output_____"
],
[
"# write to file\nif os.path.exists(crop_csv_fn):\n os.remove(crop_csv_fn)\n\n\ndf.to_csv( crop_csv_fn, header=False, index=False )",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d00105d05d5f1e74386cacf08350b830039167fb | 165,020 | ipynb | Jupyter Notebook | unsupervised ML crypto.ipynb | dmtiblin/UR-Unsupervised-Machine-Learning-Challenge | f9ffbe1113c21f93f454d629802fe0ec881ec85f | [
"ADSL"
] | null | null | null | unsupervised ML crypto.ipynb | dmtiblin/UR-Unsupervised-Machine-Learning-Challenge | f9ffbe1113c21f93f454d629802fe0ec881ec85f | [
"ADSL"
] | null | null | null | unsupervised ML crypto.ipynb | dmtiblin/UR-Unsupervised-Machine-Learning-Challenge | f9ffbe1113c21f93f454d629802fe0ec881ec85f | [
"ADSL"
] | null | null | null | 77.076133 | 40,052 | 0.693201 | [
[
[
"# Cryptocurrency Clusters",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"#import dependencies\nfrom pathlib import Path\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.manifold import TSNE\nfrom sklearn.decomposition import PCA\nfrom sklearn.cluster import KMeans",
"_____no_output_____"
]
],
[
[
"# Data Preparation",
"_____no_output_____"
]
],
[
[
"#read data in using pandas\ndf = pd.read_csv('Resources/crypto_data.csv')\ndf.head(10)\n",
"_____no_output_____"
],
[
"df.dtypes",
"_____no_output_____"
],
[
"#Discard all cryptocurrencies that are not being traded.In other words, filter for currencies that are currently being traded. \nmyfilter = (df['IsTrading'] == True)\ntrading_df = df.loc[myfilter]\ntrading_df = trading_df.drop('IsTrading', axis = 1)\ntrading_df\n#Once you have done this, drop the IsTrading column from the dataframe.",
"_____no_output_____"
],
[
"#Remove all rows that have at least one null value.\ntrading_df.dropna(how = 'any', inplace = True)\ntrading_df",
"_____no_output_____"
],
[
"#Filter for cryptocurrencies that have been mined. That is, the total coins mined should be greater than zero.\nmyfilter2 = (trading_df['TotalCoinsMined'] >0)\nfinal_df = trading_df.loc[myfilter2]\nfinal_df",
"_____no_output_____"
],
[
"#In order for your dataset to be comprehensible to a machine learning algorithm, its data should be numeric. \n#Since the coin names do not contribute to the analysis of the data, delete the CoinName from the original dataframe.\nCoinName = final_df['CoinName']\nTicker = final_df['Unnamed: 0']\nfinal_df = final_df.drop(['Unnamed: 0','CoinName'], axis = 1)\nfinal_df ",
"_____no_output_____"
],
[
"# convert the remaining features with text values, Algorithm and ProofType, into numerical data. \n#To accomplish this task, use Pandas to create dummy variables. \nfinal_df['TotalCoinSupply'] = final_df['TotalCoinSupply'].astype(float)\n",
"_____no_output_____"
],
[
"final_df = pd.get_dummies(final_df)\nfinal_df",
"_____no_output_____"
]
],
[
[
"Examine the number of rows and columns of your dataset now. How did they change?",
"_____no_output_____"
],
[
"There were 71 unique algorithms and 25 unique prooftypes so now we have 98 features in the dataset which is quite large. ",
"_____no_output_____"
]
],
[
[
"#Standardize your dataset so that columns that contain larger values do not unduly influence the outcome.\nscaled_data = StandardScaler().fit_transform(final_df)\nscaled_data",
"_____no_output_____"
]
],
[
[
"# Dimensionality Reduction ",
"_____no_output_____"
],
[
"Creating dummy variables above dramatically increased the number of features in your dataset. Perform dimensionality reduction with PCA. Rather than specify the number of principal components when you instantiate the PCA model, it is possible to state the desired explained variance.",
"_____no_output_____"
],
[
"For this project, preserve 90% of the explained variance in dimensionality reduction. \n#How did the number of the features change?",
"_____no_output_____"
]
],
[
[
"# Applying PCA to reduce dimensions\n\n# Initialize PCA model\npca = PCA(.90)\n\n# Get two principal components for the iris data.\ndata_pca = pca.fit_transform(scaled_data)",
"_____no_output_____"
],
[
"pca.explained_variance_ratio_",
"_____no_output_____"
],
[
"#df with the principal components (columns)\npd.DataFrame(data_pca)",
"_____no_output_____"
]
],
[
[
"Next, further reduce the dataset dimensions with t-SNE and visually inspect the results. In order to accomplish this task, run t-SNE on the principal components: the output of the PCA transformation. Then create a scatter plot of the t-SNE output. Observe whether there are distinct clusters or not.",
"_____no_output_____"
]
],
[
[
"# Initialize t-SNE model\ntsne = TSNE(learning_rate=35)\n# Reduce dimensions\ntsne_features = tsne.fit_transform(data_pca)",
"_____no_output_____"
],
[
"# The dataset has 2 columns\ntsne_features.shape",
"_____no_output_____"
],
[
"# Prepare to plot the dataset\n\n\n# Visualize the clusters\nplt.scatter(tsne_features[:,0], tsne_features[:,1])\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Cluster Analysis with k-Means",
"_____no_output_____"
],
[
"Create an elbow plot to identify the best number of clusters. ",
"_____no_output_____"
]
],
[
[
"#Use a for-loop to determine the inertia for each k between 1 through 10. \n#Determine, if possible, where the elbow of the plot is, and at which value of k it appears.\n\ninertia = []\nk = list(range(1, 11))\n\nfor i in k:\n km = KMeans(n_clusters=i)\n km.fit(data_pca)\n inertia.append(km.inertia_)\n \n# Define a DataFrame to plot the Elbow Curve \nelbow_data = {\"k\": k, \"inertia\": inertia}\ndf_elbow = pd.DataFrame(elbow_data)\n\nplt.plot(df_elbow['k'], df_elbow['inertia'])\nplt.xticks(range(1,11))\nplt.xlabel('Number of clusters')\nplt.ylabel('Inertia')\nplt.show()",
"_____no_output_____"
],
[
"# Initialize the K-Means model\nmodel = KMeans(n_clusters=10, random_state=0)\n\n# Train the model\nmodel.fit(scaled_data)\n\n# Predict clusters\npredictions = model.predict(scaled_data)\n\n# Create return DataFrame with predicted clusters\nfinal_df[\"cluster\"] = pd.Series(model.labels_)\n\n\nplt.figure(figsize = (18,12))\nplt.scatter(final_df['TotalCoinsMined'], final_df['TotalCoinSupply'], c=final_df['cluster'])\nplt.xlabel('TotalCoinsMined')\nplt.ylabel('TotalCoinSupply')\nplt.show()\n\nplt.figure(figsize = (18,12))\nplt.scatter(final_df['TotalCoinsMined'], final_df['TotalCoinSupply'], c=final_df['cluster'])\nplt.xlabel('TotalCoinsMined')\nplt.ylabel('TotalCoinSupply')\nplt.xlim([0, 250000000])\nplt.ylim([0, 250000000])\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Recommendation",
"_____no_output_____"
],
[
"Based on your findings, make a brief (1-2 sentences) recommendation to your clients. Can the cryptocurrencies be clustered together? If so, into how many clusters?",
"_____no_output_____"
],
[
"Even after running PCA to reduce dimensionality there are still a large number of features in the dataset. This means that there likeley was not much correlation amongst the features allowing them to be reduced together. The k-means algorithm had a very large inertia and never really leveled off, even at larger #s of clusters making it difficult to determine where an ideal # of clusters might be. In most trials, the k-means algorithm clustered most of the cryptocurrencies together in one big cluster. I would not recommend clustering the cryptocurrencies together in practice, at least not based on these data features. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d00112763ed80ce3a31e09059a2396453622c85e | 96,811 | ipynb | Jupyter Notebook | CTR Prediction/RS_Kaggle_Catboost.ipynb | amitdamri/Recommendation-Systems-Course | f8d096918b688b80c0a9acb6df3db2abe8fd8813 | [
"MIT"
] | 2 | 2021-08-23T19:15:43.000Z | 2021-11-16T13:20:04.000Z | CTR Prediction/RS_Kaggle_Catboost.ipynb | amitdamri/Recommendation-Systems-Course | f8d096918b688b80c0a9acb6df3db2abe8fd8813 | [
"MIT"
] | null | null | null | CTR Prediction/RS_Kaggle_Catboost.ipynb | amitdamri/Recommendation-Systems-Course | f8d096918b688b80c0a9acb6df3db2abe8fd8813 | [
"MIT"
] | null | null | null | 41.80095 | 2,808 | 0.403601 | [
[
[
"Our best model - Catboost with learning rate of 0.7 and 180 iterations. Was trained on 10 files of the data with similar distribution of the feature user_target_recs (among the number of rows of each feature value). We received an auc of 0.845 on the kaggle leaderboard",
"_____no_output_____"
],
[
"#Mount Drive",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount(\"/content/drive\")",
"Mounted at /content/drive\n"
]
],
[
[
"#Installations and Imports",
"_____no_output_____"
]
],
[
[
"# !pip install scikit-surprise\n!pip install catboost\n# !pip install xgboost",
"Collecting catboost\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/5a/41/24e14322b9986cf72a8763e0a0a69cc256cf963cf9502c8f0044a62c1ae8/catboost-0.26-cp37-none-manylinux1_x86_64.whl (69.2MB)\n\u001b[K |████████████████████████████████| 69.2MB 56kB/s \n\u001b[?25hRequirement already satisfied: numpy>=1.16.0 in /usr/local/lib/python3.7/dist-packages (from catboost) (1.19.5)\nRequirement already satisfied: scipy in /usr/local/lib/python3.7/dist-packages (from catboost) (1.4.1)\nRequirement already satisfied: matplotlib in /usr/local/lib/python3.7/dist-packages (from catboost) (3.2.2)\nRequirement already satisfied: graphviz in /usr/local/lib/python3.7/dist-packages (from catboost) (0.10.1)\nRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from catboost) (1.15.0)\nRequirement already satisfied: pandas>=0.24.0 in /usr/local/lib/python3.7/dist-packages (from catboost) (1.1.5)\nRequirement already satisfied: plotly in /usr/local/lib/python3.7/dist-packages (from catboost) (4.4.1)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->catboost) (1.3.1)\nRequirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->catboost) (2.8.1)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib->catboost) (0.10.0)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->catboost) (2.4.7)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.7/dist-packages (from pandas>=0.24.0->catboost) (2018.9)\nRequirement already satisfied: retrying>=1.3.3 in /usr/local/lib/python3.7/dist-packages (from plotly->catboost) (1.3.3)\nInstalling collected packages: catboost\nSuccessfully installed catboost-0.26\n"
],
[
"import os\nimport pandas as pd\n# import xgboost\n# from xgboost import XGBClassifier\n# import pickle\nimport catboost\nfrom catboost import CatBoostClassifier",
"_____no_output_____"
]
],
[
[
"#Global Parameters and Methods",
"_____no_output_____"
]
],
[
[
"home_path = \"/content/drive/MyDrive/RS_Kaggle_Competition\"",
"_____no_output_____"
],
[
"\ndef get_train_files_paths(path):\n dir_paths = [ os.path.join(path, dir_name) for dir_name in os.listdir(path) if dir_name.startswith(\"train\")]\n file_paths = []\n\n for dir_path in dir_paths:\n curr_dir_file_paths = [ os.path.join(dir_path, file_name) for file_name in os.listdir(dir_path) ]\n file_paths.extend(curr_dir_file_paths)\n \n return file_paths\n\ntrain_file_paths = get_train_files_paths(home_path)",
"_____no_output_____"
]
],
[
[
"#Get Data",
"_____no_output_____"
]
],
[
[
"def get_df_of_many_files(paths_list, number_of_files):\n for i in range(number_of_files):\n path = paths_list[i]\n curr_df = pd.read_csv(path)\n\n if i == 0:\n df = curr_df\n else:\n df = pd.concat([df, curr_df])\n \n return df\n",
"_____no_output_____"
],
[
"sample_train_data = get_df_of_many_files(train_file_paths[-21:], 10)\n# sample_train_data = pd.read_csv(home_path + \"/10_files_train_data\")",
"_____no_output_____"
],
[
"sample_val_data = get_df_of_many_files(train_file_paths[-10:], 3)\n# sample_val_data = pd.read_csv(home_path+\"/3_files_val_data\")",
"_____no_output_____"
],
[
"# sample_val_data.to_csv(home_path+\"/3_files_val_data\")",
"_____no_output_____"
]
],
[
[
"#Preprocess data",
"_____no_output_____"
]
],
[
[
"train_data = sample_train_data.fillna(\"Unknown\")\n",
"_____no_output_____"
],
[
"val_data = sample_val_data.fillna(\"Unknown\")\n",
"_____no_output_____"
],
[
"train_data",
"_____no_output_____"
],
[
"import gc\n\ndel sample_val_data\ndel sample_train_data\ngc.collect()",
"_____no_output_____"
]
],
[
[
"## Scale columns",
"_____no_output_____"
]
],
[
[
"# scale columns\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.preprocessing import MinMaxScaler\n\nscaling_cols= [\"empiric_calibrated_recs\", \"empiric_clicks\", \"empiric_calibrated_recs\", \"user_recs\", \"user_clicks\", \"user_target_recs\"]\n\nscaler = StandardScaler()\n\ntrain_data[scaling_cols] = scaler.fit_transform(train_data[scaling_cols])\n\nval_data[scaling_cols] = scaler.transform(val_data[scaling_cols])",
"_____no_output_____"
],
[
"train_data",
"_____no_output_____"
],
[
"val_data = val_data.drop(columns=[\"Unnamed: 0.1\"])\nval_data",
"_____no_output_____"
]
],
[
[
"#Explore Data",
"_____no_output_____"
]
],
[
[
"sample_train_data",
"_____no_output_____"
],
[
"test_data",
"_____no_output_____"
],
[
"from collections import Counter\nuser_recs_dist = test_data[\"user_recs\"].value_counts(normalize=True)\n\ntop_user_recs_count = user_recs_dist.nlargest(200)\n\nprint(top_user_recs_count)\n\npercent = sum(top_user_recs_count.values)\npercent",
"_____no_output_____"
],
[
"print(sample_train_data[\"user_recs\"].value_counts(normalize=False))\nprint(test_data[\"user_recs\"].value_counts())",
"0.0 542238\n300.0 50285\n3.0 46951\n1.0 45815\n2.0 39176\n ... \n3262.0 1\n3795.0 1\n3793.0 1\n3788.0 1\n3224.0 1\nName: user_recs, Length: 3947, dtype: int64\n0.0 112851\n3.0 10185\n1.0 10088\n2.0 8581\n4.0 6510\n ... \n2190.0 1\n2549.0 1\n2720.0 1\n2289.0 1\n2588.0 1\nName: user_recs, Length: 2534, dtype: int64\n"
],
[
"positions = top_user_recs_count\ndef sample(obj, replace=False, total=1500000):\n return obj.sample(n=int(positions[obj.name] * total), replace=replace)\n\nsample_train_data_filtered = sample_train_data[sample_train_data[\"user_recs\"].isin(positions.keys())]\nresult = sample_train_data_filtered.groupby(\"user_recs\").apply(sample).reset_index(drop=True)\nresult[\"user_recs\"].value_counts(normalize=True)",
"_____no_output_____"
],
[
"top_user_recs_train_data = result",
"_____no_output_____"
],
[
"top_user_recs_train_data",
"_____no_output_____"
],
[
"not_top_user_recs_train_data = sample_train_data[~sample_train_data[\"user_recs\"].isin(top_user_recs_train_data[\"user_recs\"].unique())]\nnot_top_user_recs_train_data[\"user_recs\"].value_counts()",
"_____no_output_____"
],
[
"train_data = pd.concat([top_user_recs_train_data, not_top_user_recs_train_data])",
"_____no_output_____"
],
[
"train_data[\"user_recs\"].value_counts(normalize=False)",
"_____no_output_____"
],
[
"train_data = train_data.drop(columns = [\"user_id_hash\"])",
"_____no_output_____"
],
[
"train_data = train_data.fillna(\"Unknown\")",
"_____no_output_____"
],
[
"train_data",
"_____no_output_____"
]
],
[
[
"#Train the model",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score\nfrom sklearn import metrics\n\n\nX_train = train_data.drop(columns=[\"is_click\"], inplace=False)\ny_train = train_data[\"is_click\"]\n\nX_val = val_data.drop(columns=[\"is_click\"], inplace=False)\ny_val = val_data[\"is_click\"]",
"_____no_output_____"
],
[
"from catboost import CatBoostClassifier\n# cat_features_inds = [1,2,3,4,7,8,12,13,14,15,17,18]\nencode_cols = [ \"user_id_hash\", \"target_id_hash\", \"syndicator_id_hash\", \"campaign_id_hash\", \"target_item_taxonomy\", \"placement_id_hash\", \"publisher_id_hash\", \"source_id_hash\", \"source_item_type\", \"browser_platform\", \"country_code\", \"region\", \"gmt_offset\"]\n\n# model = CatBoostClassifier(iterations = 50, learning_rate=0.5, task_type='CPU', loss_function='Logloss', cat_features = encode_cols)\nmodel = CatBoostClassifier(iterations = 180, learning_rate=0.7, task_type='CPU', loss_function='Logloss', cat_features = encode_cols,\n eval_metric='AUC')#, depth=6, l2_leaf_reg= 10)\n\n\"\"\"\nAll of our tries with catboost (only the best of them were uploaded to kaggle):\n\nresults:\nall features, all rows of train fillna = Unknown\nlogloss 100 iterations learning rate 0.5 10 files: 0.857136889762303 | bestTest = 0.4671640673 0.857136889762303\nlogloss 100 iterations learning rate 0.4 10 files: bestTest = 0.4676805926 0.856750110976787\n\nlogloss 100 iterations learning rate 0.55 10 files: bestTest = 0.4669830858 0.8572464626142212\n\nlogloss 120 iterations learning rate 0.6 10 files: bestTest = 0.4662084678 0.8577564702279399\n\nlogloss 150 iterations learning rate 0.7 10 files: bestTest = 0.4655981391 0.8581645278496352\n\nlogloss 180 iterations learning rate 0.7 10 files: bestTest = 0.4653168207 0.8583423138228865 !!!!!!!!!!\n\nlogloss 180 iterations learning rate 0.7 10 files day extracted from date (not as categorical): 0.8583034988\n\nlogloss 180 iterations learning rate 0.7 10 files day extracted from date (as categorical): 0.8583014151\n\nlogloss 180 iterations learning rate 0.75 10 files day extracted from date (as categorical): 0.8582889749\n\nlogloss 180 iterations learning rate 0.65 10 files day extracted from date (as categorical): 0.8582334254\n\nlogloss 180 iterations learning rate 0.65 10 files day extracted from date (as categorical) StandardScaler: 0.8582101013\n\nlogloss 180 iterations learning rate 0.7 10 files day extracted from date (as categorical) MinMaxScaler dropna: ~0.8582\n\nlogloss 180 iterations learning rate 0.7 distributed data train and val, day extracted as categorical MinMaxScaler: 0.8561707 \n\nlogloss 180 iterations learning rate 0.7 distributed data train and val, day extracted as not categorical no scale: 0.8561707195\n\nlogloss 180 iterations learning rate 0.7 distributed data train and val, no scale no date: 0.8559952294\n\nlogloss 180 iterations learning rate 0.7 distributed data train and val, day extracted as not categorical no scale with date: 0.8560461554\n\nlogloss 180 iterations learning rate 0.7, 9 times distributed data train and val, no user no date: 0.8545560094\n\nlogloss 180 iterations learning rate 0.7, 9 times distributed data train and val, with user and numeric day: 0.8561601034\n\nlogloss 180 iterations learning rate 0.7, 9 times distributed data train and val, with user with numeric date: 0.8568834122\n\nlogloss 180 iterations learning rate 0.7, 10 different files, scaled, all features: 0.8584829166 !!!!!!!\n\nlogloss 180 iterations learning rate 0.7, new data, scaled, all features: 0.8915972905 test: 0.84108\n\n\nlogloss 180 iterations learning rate 0.9 10 files: bestTest = 0.4656462845\n\nlogloss 100 iterations learning rate 0.5 8 files: 0.8568031111965864\nlogloss 300 iterations learning rate 0.5: \ncrossentropy 50 iterations learning rate 0.5: 0.8556282878645277\n\"\"\"",
"_____no_output_____"
],
[
"model.fit(X_train, y_train, eval_set=(X_val, y_val), verbose=10)",
"0:\ttest: 0.8149026\tbest: 0.8149026 (0)\ttotal: 6.36s\tremaining: 18m 57s\n10:\ttest: 0.8461028\tbest: 0.8461028 (10)\ttotal: 53.6s\tremaining: 13m 44s\n20:\ttest: 0.8490288\tbest: 0.8490288 (20)\ttotal: 1m 38s\tremaining: 12m 26s\n30:\ttest: 0.8505695\tbest: 0.8505695 (30)\ttotal: 2m 23s\tremaining: 11m 29s\n40:\ttest: 0.8514950\tbest: 0.8514950 (40)\ttotal: 3m 8s\tremaining: 10m 38s\n50:\ttest: 0.8522340\tbest: 0.8522340 (50)\ttotal: 3m 53s\tremaining: 9m 50s\n60:\ttest: 0.8526374\tbest: 0.8526374 (60)\ttotal: 4m 37s\tremaining: 9m\n70:\ttest: 0.8531463\tbest: 0.8531463 (70)\ttotal: 5m 22s\tremaining: 8m 14s\n80:\ttest: 0.8534035\tbest: 0.8534035 (80)\ttotal: 6m 6s\tremaining: 7m 27s\n90:\ttest: 0.8536159\tbest: 0.8536567 (89)\ttotal: 6m 51s\tremaining: 6m 42s\n100:\ttest: 0.8537674\tbest: 0.8537674 (100)\ttotal: 7m 35s\tremaining: 5m 56s\n110:\ttest: 0.8539636\tbest: 0.8539636 (110)\ttotal: 8m 19s\tremaining: 5m 10s\n120:\ttest: 0.8541628\tbest: 0.8541628 (120)\ttotal: 9m 3s\tremaining: 4m 25s\n130:\ttest: 0.8542642\tbest: 0.8542642 (130)\ttotal: 9m 48s\tremaining: 3m 39s\n140:\ttest: 0.8543702\tbest: 0.8543800 (137)\ttotal: 10m 31s\tremaining: 2m 54s\n150:\ttest: 0.8544469\tbest: 0.8544550 (149)\ttotal: 11m 15s\tremaining: 2m 9s\n160:\ttest: 0.8543904\tbest: 0.8545011 (158)\ttotal: 11m 59s\tremaining: 1m 24s\n170:\ttest: 0.8543992\tbest: 0.8545011 (158)\ttotal: 12m 43s\tremaining: 40.2s\n179:\ttest: 0.8544623\tbest: 0.8545011 (158)\ttotal: 13m 23s\tremaining: 0us\n\nbestTest = 0.8545011269\nbestIteration = 158\n\nShrink model to first 159 iterations.\n"
]
],
[
[
"# Submission File",
"_____no_output_____"
]
],
[
[
"test_data = pd.read_csv(\"/content/drive/MyDrive/RS_Kaggle_Competition/test/test_file.csv\")\n",
"_____no_output_____"
],
[
"test_data = test_data.iloc[:,:-1]",
"_____no_output_____"
],
[
"test_data[scaling_cols] = scaler.transform(test_data[scaling_cols])",
"_____no_output_____"
],
[
"\nX_test = test_data.fillna(\"Unknown\")\nX_test",
"_____no_output_____"
],
[
"pred_proba = model.predict_proba(X_test)\n\nsubmission_dir_path = \"/content/drive/MyDrive/RS_Kaggle_Competition/submissions\"\npred = pred_proba[:,1]\n\npred_df = pd.DataFrame(pred)\npred_df.reset_index(inplace=True)\npred_df.columns = ['Id', 'Predicted']\npred_df.to_csv(submission_dir_path + '/catboost_submission_datafrom1704_data_lr_0.7_with_scale_with_num_startdate_with_user_iters_159.csv', index=False)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d001138d46ac9adce3ad006c5b68ae3c9b8221ce | 161,559 | ipynb | Jupyter Notebook | Randomized Optimization/NQueens.ipynb | cindynyoumsigit/MachineLearning | 383fb849ac1e98c2e96e7f0f241ba57fb99ed956 | [
"Apache-2.0"
] | null | null | null | Randomized Optimization/NQueens.ipynb | cindynyoumsigit/MachineLearning | 383fb849ac1e98c2e96e7f0f241ba57fb99ed956 | [
"Apache-2.0"
] | null | null | null | Randomized Optimization/NQueens.ipynb | cindynyoumsigit/MachineLearning | 383fb849ac1e98c2e96e7f0f241ba57fb99ed956 | [
"Apache-2.0"
] | null | null | null | 175.990196 | 50,152 | 0.871688 | [
[
[
"# Random Search Algorithms",
"_____no_output_____"
],
[
"### Importing Necessary Libraries\n",
"_____no_output_____"
]
],
[
[
"import six\nimport sys\nsys.modules['sklearn.externals.six'] = six\nimport mlrose\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport mlrose_hiive\nimport matplotlib.pyplot as plt\nnp.random.seed(44)\nsns.set_style(\"darkgrid\")",
"_____no_output_____"
]
],
[
[
"### Defining a Fitness Function Object ",
"_____no_output_____"
]
],
[
[
"# Define alternative N-Queens fitness function for maximization problem\ndef queens_max(state):\n \n # Initialize counter\n fitness = 0\n \n # For all pairs of queens\n for i in range(len(state) - 1):\n for j in range(i + 1, len(state)):\n \n # Check for horizontal, diagonal-up and diagonal-down attacks\n if (state[j] != state[i]) \\\n and (state[j] != state[i] + (j - i)) \\\n and (state[j] != state[i] - (j - i)):\n \n # If no attacks, then increment counter\n fitness += 1\n\n return fitness\n\n# Initialize custom fitness function object\nfitness_cust = mlrose.CustomFitness(queens_max)",
"_____no_output_____"
]
],
[
[
"### Defining an Optimization Problem Object",
"_____no_output_____"
]
],
[
[
"%%time\n# DiscreteOpt() takes integers in range 0 to max_val -1 defined at initialization\nnumber_of_queens = 16\nproblem = mlrose_hiive.DiscreteOpt(length = number_of_queens, fitness_fn = fitness_cust, maximize = True, max_val = number_of_queens)",
"CPU times: user 138 µs, sys: 79 µs, total: 217 µs\nWall time: 209 µs\n"
]
],
[
[
"### Optimization #1 Simulated Annealing",
"_____no_output_____"
]
],
[
[
"%%time\nsa = mlrose_hiive.SARunner(problem, experiment_name=\"SA_Exp\", \n iteration_list=[10000],\n temperature_list=[10, 50, 100, 250, 500],\n decay_list=[mlrose_hiive.ExpDecay,\n mlrose_hiive.GeomDecay],\n seed=44, max_attempts=100)\n\nsa_run_stats, sa_run_curves = sa.run()",
"CPU times: user 13.6 s, sys: 103 ms, total: 13.7 s\nWall time: 13.6 s\n"
],
[
"last_iters = sa_run_stats[sa_run_stats.Iteration != 0].reset_index()\nprint('Mean:', last_iters.Fitness.mean(), '\\nMin:',last_iters.Fitness.max(),'\\nMax:',last_iters.Fitness.max())\nprint('Mean Time;',last_iters.Time.mean())",
"Mean: 118.7 \nMin: 119.0 \nMax: 119.0\nMean Time; 1.3441894618999999\n"
],
[
"best_index_in_curve = sa_run_curves.Fitness.idxmax()\nbest_decay = sa_run_curves.iloc[best_index_in_curve].Temperature\nbest_curve = sa_run_curves.loc[sa_run_curves.Temperature == best_decay, :]\nbest_curve.reset_index(inplace=True)\nbest_decay",
"_____no_output_____"
],
[
"best_index_in_curve = sa_run_curves.Fitness.idxmax()\nbest_decay = sa_run_curves.iloc[best_index_in_curve].Temperature\nbest_sa_curve = sa_run_curves.loc[sa_run_curves.Temperature == best_decay, :]\nbest_sa_curve.reset_index(inplace=True)\n\n\n# draw lineplot \nsa_run_curves['Temperature'] = sa_run_curves['Temperature'].astype(str).astype(float)\nsa_run_curves_t1 = sa_run_curves[sa_run_curves['Temperature'] == 10] \nsa_run_curves_t2 = sa_run_curves[sa_run_curves['Temperature'] == 50] \nsa_run_curves_t3 = sa_run_curves[sa_run_curves['Temperature'] == 100]\nsa_run_curves_t4 = sa_run_curves[sa_run_curves['Temperature'] == 250] \nsa_run_curves_t5 = sa_run_curves[sa_run_curves['Temperature'] == 500] \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=sa_run_curves_t1, label = \"t = 10\")\nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=sa_run_curves_t2, label = \"t = 50\") \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=sa_run_curves_t3, label = \"t = 100\") \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=sa_run_curves_t4, label = \"t = 250\") \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=sa_run_curves_t5, label = \"t = 500\") \n \nplt.title('16-Queens SA Fitness Vs Iterations')\nplt.show()\n\nsa_run_curves",
"_____no_output_____"
]
],
[
[
"### Optimization #2 Genetic Algorithm",
"_____no_output_____"
]
],
[
[
"%%time\nga = mlrose_hiive.GARunner(problem=problem,\n experiment_name=\"GA_Exp\",\n seed=44,\n iteration_list = [10000],\n max_attempts = 100,\n population_sizes = [100, 500],\n mutation_rates = [0.1, 0.25, 0.5])\n\nga_run_stats, ga_run_curves = ga.run()",
"CPU times: user 1min 10s, sys: 161 ms, total: 1min 10s\nWall time: 1min 11s\n"
],
[
"last_iters = ga_run_stats[ga_run_stats.Iteration != 0].reset_index()\nprint(\"Max and mean\")\nprint(last_iters.Fitness.max(), last_iters.Fitness.mean(), last_iters.Time.mean())\nprint(last_iters.groupby(\"Mutation Rate\").Fitness.mean())\nprint(last_iters.groupby(\"Population Size\").Fitness.mean())\nprint(last_iters.groupby(\"Population Size\").Time.mean())",
"Max and mean\n120.0 118.66666666666667 11.860036887000001\nMutation Rate\n0.10 119.5\n0.25 118.5\n0.50 118.0\nName: Fitness, dtype: float64\nPopulation Size\n100 118.000000\n500 119.333333\nName: Fitness, dtype: float64\nPopulation Size\n100 2.803024\n500 20.917050\nName: Time, dtype: float64\n"
],
[
"# draw lineplot \nga_run_curves_mu1 = ga_run_curves[ga_run_curves['Mutation Rate'] == 0.1] \nga_run_curves_mu2 = ga_run_curves[ga_run_curves['Mutation Rate'] == 0.25] \nga_run_curves_mu3 = ga_run_curves[ga_run_curves['Mutation Rate'] == 0.5] \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=ga_run_curves_mu1, label = \"mr = 0.1\") \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=ga_run_curves_mu2, label = \"mr = 0.25\") \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=ga_run_curves_mu3, label = \"mr = 0.5\") \nplt.title('16-Queens GA Fitness Vs Iterations')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Optimization #3 MIMIC",
"_____no_output_____"
]
],
[
[
"%%time\nmmc = mlrose_hiive.MIMICRunner(problem=problem,\n experiment_name=\"MMC_Exp\",\n seed=44,\n iteration_list=[10000],\n max_attempts=100,\n population_sizes=[100,500],\n keep_percent_list=[0.1, 0.25, 0.5],\n use_fast_mimic=True)\n\n# the two data frames will contain the results\nmmc_run_stats, mmc_run_curves = mmc.run()",
"CPU times: user 56.8 s, sys: 726 ms, total: 57.5 s\nWall time: 57.8 s\n"
],
[
"last_iters = mmc_run_stats[mmc_run_stats.Iteration != 0].reset_index()\nprint(\"Max and mean\")\nprint(last_iters.Fitness.max(), last_iters.Fitness.mean(), last_iters.Time.mean())\nprint(last_iters.groupby(\"Keep Percent\").Fitness.mean())\nprint(last_iters.groupby(\"Population Size\").Fitness.mean())\nprint(last_iters.groupby(\"Population Size\").Time.mean())\nmmc_run_curves",
"Max and mean\n119.0 115.33333333333333 9.633991666500004\nKeep Percent\n0.10 115.0\n0.25 116.0\n0.50 115.0\nName: Fitness, dtype: float64\nPopulation Size\n100 113.666667\n500 117.000000\nName: Fitness, dtype: float64\nPopulation Size\n100 4.481067\n500 14.786917\nName: Time, dtype: float64\n"
],
[
"# draw lineplot \nmmc_run_curves_kp1 = mmc_run_curves[mmc_run_curves['Keep Percent'] == 0.1] \nmmc_run_curves_kp2 = mmc_run_curves[mmc_run_curves['Keep Percent'] == 0.25] \nmmc_run_curves_kp3 = mmc_run_curves[mmc_run_curves['Keep Percent'] == 0.5] \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=mmc_run_curves_kp1, label = \"kp = 0.1\") \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=mmc_run_curves_kp2, label = \"kp = 0.25\") \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=mmc_run_curves_kp3, label = \"kp = 0.5\") \nplt.title('16-Queens MIMIC Fitness Vs Iterations')\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### Optimization #4 Randomized Hill Climbing\n",
"_____no_output_____"
]
],
[
[
"%%time\nrunner_return = mlrose_hiive.RHCRunner(problem, experiment_name=\"first_try\", \n iteration_list=[10000],\n seed=44, max_attempts=100, \n restart_list=[100])\nrhc_run_stats, rhc_run_curves = runner_return.run()",
"CPU times: user 3min 12s, sys: 362 ms, total: 3min 12s\nWall time: 3min 12s\n"
],
[
"last_iters = rhc_run_stats[rhc_run_stats.Iteration != 0].reset_index()\nprint(last_iters.Fitness.mean(), last_iters.Fitness.max())\nprint(last_iters.Time.max())",
"116.78217821782178 119.0\n192.82425359899997\n"
],
[
"best_index_in_curve = rhc_run_curves.Fitness.idxmax()\nbest_decay = rhc_run_curves.iloc[best_index_in_curve].current_restart\nbest_RHC_curve = rhc_run_curves.loc[rhc_run_curves.current_restart == best_decay, :]\nbest_RHC_curve.reset_index(inplace=True)\nbest_RHC_curve\n# draw lineplot \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=best_RHC_curve) \nplt.title('16-Queens RHC Fitness Vs Iterations')\nplt.show()",
"_____no_output_____"
],
[
"sns.lineplot(x=\"Iteration\", y=\"Fitness\", data=ga_run_curves_mu3, label = \"GA\") \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=best_sa_curve, label = \"SA\") \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=mmc_run_curves, label = \"MIMIC\") \nsns.lineplot(x=\"Iteration\", y=\"Fitness\", data=best_RHC_curve, label = \"RHC\") \nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0011b0973cc06f30ce5fa93f30b51deaaf3b933 | 21,954 | ipynb | Jupyter Notebook | docs/_downloads/a584d8a4ce8e691ca795984e7a5eedbd/tuning_guide.ipynb | junhyung9985/PyTorch-tutorials-kr | 07c50e5ddfc2f118f01ecbc071a24763f9891171 | [
"BSD-3-Clause"
] | null | null | null | docs/_downloads/a584d8a4ce8e691ca795984e7a5eedbd/tuning_guide.ipynb | junhyung9985/PyTorch-tutorials-kr | 07c50e5ddfc2f118f01ecbc071a24763f9891171 | [
"BSD-3-Clause"
] | null | null | null | docs/_downloads/a584d8a4ce8e691ca795984e7a5eedbd/tuning_guide.ipynb | junhyung9985/PyTorch-tutorials-kr | 07c50e5ddfc2f118f01ecbc071a24763f9891171 | [
"BSD-3-Clause"
] | null | null | null | 88.524194 | 1,542 | 0.686435 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\nPerformance Tuning Guide\n*************************\n**Author**: `Szymon Migacz <https://github.com/szmigacz>`_\n\nPerformance Tuning Guide is a set of optimizations and best practices which can\naccelerate training and inference of deep learning models in PyTorch. Presented\ntechniques often can be implemented by changing only a few lines of code and can\nbe applied to a wide range of deep learning models across all domains.\n\nGeneral optimizations\n---------------------\n",
"_____no_output_____"
],
[
"Enable async data loading and augmentation\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n`torch.utils.data.DataLoader <https://pytorch.org/docs/stable/data.html#torch.utils.data.DataLoader>`_\nsupports asynchronous data loading and data augmentation in separate worker\nsubprocesses. The default setting for ``DataLoader`` is ``num_workers=0``,\nwhich means that the data loading is synchronous and done in the main process.\nAs a result the main training process has to wait for the data to be available\nto continue the execution.\n\nSetting ``num_workers > 0`` enables asynchronous data loading and overlap\nbetween the training and data loading. ``num_workers`` should be tuned\ndepending on the workload, CPU, GPU, and location of training data.\n\n``DataLoader`` accepts ``pin_memory`` argument, which defaults to ``False``.\nWhen using a GPU it's better to set ``pin_memory=True``, this instructs\n``DataLoader`` to use pinned memory and enables faster and asynchronous memory\ncopy from the host to the GPU.\n\n",
"_____no_output_____"
],
[
"Disable gradient calculation for validation or inference\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\nPyTorch saves intermediate buffers from all operations which involve tensors\nthat require gradients. Typically gradients aren't needed for validation or\ninference.\n`torch.no_grad() <https://pytorch.org/docs/stable/generated/torch.no_grad.html#torch.no_grad>`_\ncontext manager can be applied to disable gradient calculation within a\nspecified block of code, this accelerates execution and reduces the amount of\nrequired memory.\n`torch.no_grad() <https://pytorch.org/docs/stable/generated/torch.no_grad.html#torch.no_grad>`_\ncan also be used as a function decorator.\n\n",
"_____no_output_____"
],
[
"Disable bias for convolutions directly followed by a batch norm\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n`torch.nn.Conv2d() <https://pytorch.org/docs/stable/generated/torch.nn.Conv2d.html#torch.nn.Conv2d>`_\nhas ``bias`` parameter which defaults to ``True`` (the same is true for\n`Conv1d <https://pytorch.org/docs/stable/generated/torch.nn.Conv1d.html#torch.nn.Conv1d>`_\nand\n`Conv3d <https://pytorch.org/docs/stable/generated/torch.nn.Conv3d.html#torch.nn.Conv3d>`_\n).\n\nIf a ``nn.Conv2d`` layer is directly followed by a ``nn.BatchNorm2d`` layer,\nthen the bias in the convolution is not needed, instead use\n``nn.Conv2d(..., bias=False, ....)``. Bias is not needed because in the first\nstep ``BatchNorm`` subtracts the mean, which effectively cancels out the\neffect of bias.\n\nThis is also applicable to 1d and 3d convolutions as long as ``BatchNorm`` (or\nother normalization layer) normalizes on the same dimension as convolution's\nbias.\n\nModels available from `torchvision <https://github.com/pytorch/vision>`_\nalready implement this optimization.\n\n",
"_____no_output_____"
],
[
"Use parameter.grad = None instead of model.zero_grad() or optimizer.zero_grad()\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\nInstead of calling:\n\n",
"_____no_output_____"
]
],
[
[
"model.zero_grad()\n# or\noptimizer.zero_grad()",
"_____no_output_____"
]
],
[
[
"to zero out gradients, use the following method instead:\n\n",
"_____no_output_____"
]
],
[
[
"for param in model.parameters():\n param.grad = None",
"_____no_output_____"
]
],
[
[
"The second code snippet does not zero the memory of each individual parameter,\nalso the subsequent backward pass uses assignment instead of addition to store\ngradients, this reduces the number of memory operations.\n\nSetting gradient to ``None`` has a slightly different numerical behavior than\nsetting it to zero, for more details refer to the\n`documentation <https://pytorch.org/docs/master/optim.html#torch.optim.Optimizer.zero_grad>`_.\n\nAlternatively, starting from PyTorch 1.7, call ``model`` or\n``optimizer.zero_grad(set_to_none=True)``.\n\n",
"_____no_output_____"
],
[
"Fuse pointwise operations\n~~~~~~~~~~~~~~~~~~~~~~~~~\nPointwise operations (elementwise addition, multiplication, math functions -\n``sin()``, ``cos()``, ``sigmoid()`` etc.) can be fused into a single kernel\nto amortize memory access time and kernel launch time.\n\n`PyTorch JIT <https://pytorch.org/docs/stable/jit.html>`_ can fuse kernels\nautomatically, although there could be additional fusion opportunities not yet\nimplemented in the compiler, and not all device types are supported equally.\n\nPointwise operations are memory-bound, for each operation PyTorch launches a\nseparate kernel. Each kernel loads data from the memory, performs computation\n(this step is usually inexpensive) and stores results back into the memory.\n\nFused operator launches only one kernel for multiple fused pointwise ops and\nloads/stores data only once to the memory. This makes JIT very useful for\nactivation functions, optimizers, custom RNN cells etc.\n\nIn the simplest case fusion can be enabled by applying\n`torch.jit.script <https://pytorch.org/docs/stable/generated/torch.jit.script.html#torch.jit.script>`_\ndecorator to the function definition, for example:\n\n",
"_____no_output_____"
]
],
[
[
"@torch.jit.script\ndef fused_gelu(x):\n return x * 0.5 * (1.0 + torch.erf(x / 1.41421))",
"_____no_output_____"
]
],
[
[
"Refer to\n`TorchScript documentation <https://pytorch.org/docs/stable/jit.html>`_\nfor more advanced use cases.\n\n",
"_____no_output_____"
],
[
"Enable channels_last memory format for computer vision models\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\nPyTorch 1.5 introduced support for ``channels_last`` memory format for\nconvolutional networks. This format is meant to be used in conjunction with\n`AMP <https://pytorch.org/docs/stable/amp.html>`_ to further accelerate\nconvolutional neural networks with\n`Tensor Cores <https://www.nvidia.com/en-us/data-center/tensor-cores/>`_.\n\nSupport for ``channels_last`` is experimental, but it's expected to work for\nstandard computer vision models (e.g. ResNet-50, SSD). To convert models to\n``channels_last`` format follow\n`Channels Last Memory Format Tutorial <https://tutorials.pytorch.kr/intermediate/memory_format_tutorial.html>`_.\nThe tutorial includes a section on\n`converting existing models <https://tutorials.pytorch.kr/intermediate/memory_format_tutorial.html#converting-existing-models>`_.\n\n",
"_____no_output_____"
],
[
"Checkpoint intermediate buffers\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\nBuffer checkpointing is a technique to mitigate the memory capacity burden of\nmodel training. Instead of storing inputs of all layers to compute upstream\ngradients in backward propagation, it stores the inputs of a few layers and\nthe others are recomputed during backward pass. The reduced memory\nrequirements enables increasing the batch size that can improve utilization.\n\nCheckpointing targets should be selected carefully. The best is not to store\nlarge layer outputs that have small re-computation cost. The example target\nlayers are activation functions (e.g. ``ReLU``, ``Sigmoid``, ``Tanh``),\nup/down sampling and matrix-vector operations with small accumulation depth.\n\nPyTorch supports a native\n`torch.utils.checkpoint <https://pytorch.org/docs/stable/checkpoint.html>`_\nAPI to automatically perform checkpointing and recomputation.\n\n",
"_____no_output_____"
],
[
"Disable debugging APIs\n~~~~~~~~~~~~~~~~~~~~~~\nMany PyTorch APIs are intended for debugging and should be disabled for\nregular training runs:\n\n* anomaly detection:\n `torch.autograd.detect_anomaly <https://pytorch.org/docs/stable/autograd.html#torch.autograd.detect_anomaly>`_\n or\n `torch.autograd.set_detect_anomaly(True) <https://pytorch.org/docs/stable/autograd.html#torch.autograd.set_detect_anomaly>`_\n* profiler related:\n `torch.autograd.profiler.emit_nvtx <https://pytorch.org/docs/stable/autograd.html#torch.autograd.profiler.emit_nvtx>`_,\n `torch.autograd.profiler.profile <https://pytorch.org/docs/stable/autograd.html#torch.autograd.profiler.profile>`_\n* autograd gradcheck:\n `torch.autograd.gradcheck <https://pytorch.org/docs/stable/autograd.html#torch.autograd.gradcheck>`_\n or\n `torch.autograd.gradgradcheck <https://pytorch.org/docs/stable/autograd.html#torch.autograd.gradgradcheck>`_\n\n\n",
"_____no_output_____"
],
[
"GPU specific optimizations\n--------------------------\n\n",
"_____no_output_____"
],
[
"Enable cuDNN auto-tuner\n~~~~~~~~~~~~~~~~~~~~~~~\n`NVIDIA cuDNN <https://developer.nvidia.com/cudnn>`_ supports many algorithms\nto compute a convolution. Autotuner runs a short benchmark and selects the\nkernel with the best performance on a given hardware for a given input size.\n\nFor convolutional networks (other types currently not supported), enable cuDNN\nautotuner before launching the training loop by setting:\n\n",
"_____no_output_____"
]
],
[
[
"torch.backends.cudnn.benchmark = True",
"_____no_output_____"
]
],
[
[
"* the auto-tuner decisions may be non-deterministic; different algorithm may\n be selected for different runs. For more details see\n `PyTorch: Reproducibility <https://pytorch.org/docs/stable/notes/randomness.html?highlight=determinism>`_\n* in some rare cases, such as with highly variable input sizes, it's better\n to run convolutional networks with autotuner disabled to avoid the overhead\n associated with algorithm selection for each input size.\n\n\n",
"_____no_output_____"
],
[
"Avoid unnecessary CPU-GPU synchronization\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\nAvoid unnecessary synchronizations, to let the CPU run ahead of the\naccelerator as much as possible to make sure that the accelerator work queue\ncontains many operations.\n\nWhen possible, avoid operations which require synchronizations, for example:\n\n* ``print(cuda_tensor)``\n* ``cuda_tensor.item()``\n* memory copies: ``tensor.cuda()``, ``cuda_tensor.cpu()`` and equivalent\n ``tensor.to(device)`` calls\n* ``cuda_tensor.nonzero()``\n* python control flow which depends on results of operations performed on cuda\n tensors e.g. ``if (cuda_tensor != 0).all()``\n\n\n",
"_____no_output_____"
],
[
"Create tensors directly on the target device\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\nInstead of calling ``torch.rand(size).cuda()`` to generate a random tensor,\nproduce the output directly on the target device:\n``torch.rand(size, device=torch.device('cuda'))``.\n\nThis is applicable to all functions which create new tensors and accept\n``device`` argument:\n`torch.rand() <https://pytorch.org/docs/stable/generated/torch.rand.html#torch.rand>`_,\n`torch.zeros() <https://pytorch.org/docs/stable/generated/torch.zeros.html#torch.zeros>`_,\n`torch.full() <https://pytorch.org/docs/stable/generated/torch.full.html#torch.full>`_\nand similar.\n\n",
"_____no_output_____"
],
[
"Use mixed precision and AMP\n~~~~~~~~~~~~~~~~~~~~~~~~~~~\nMixed precision leverages\n`Tensor Cores <https://www.nvidia.com/en-us/data-center/tensor-cores/>`_\nand offers up to 3x overall speedup on Volta and newer GPU architectures. To\nuse Tensor Cores AMP should be enabled and matrix/tensor dimensions should\nsatisfy requirements for calling kernels that use Tensor Cores.\n\nTo use Tensor Cores:\n\n* set sizes to multiples of 8 (to map onto dimensions of Tensor Cores)\n\n * see\n `Deep Learning Performance Documentation\n <https://docs.nvidia.com/deeplearning/performance/index.html#optimizing-performance>`_\n for more details and guidelines specific to layer type\n * if layer size is derived from other parameters rather than fixed, it can\n still be explicitly padded e.g. vocabulary size in NLP models\n\n* enable AMP\n\n * Introduction to Mixed Precision Training and AMP:\n `video <https://www.youtube.com/watch?v=jF4-_ZK_tyc&feature=youtu.be>`_,\n `slides <https://nvlabs.github.io/eccv2020-mixed-precision-tutorial/files/dusan_stosic-training-neural-networks-with-tensor-cores.pdf>`_\n * native PyTorch AMP is available starting from PyTorch 1.6:\n `documentation <https://pytorch.org/docs/stable/amp.html>`_,\n `examples <https://pytorch.org/docs/stable/notes/amp_examples.html#amp-examples>`_,\n `tutorial <https://tutorials.pytorch.kr/recipes/recipes/amp_recipe.html>`_\n\n\n\n",
"_____no_output_____"
],
[
"Pre-allocate memory in case of variable input length\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\nModels for speech recognition or for NLP are often trained on input tensors\nwith variable sequence length. Variable length can be problematic for PyTorch\ncaching allocator and can lead to reduced performance or to unexpected\nout-of-memory errors. If a batch with a short sequence length is followed by\nan another batch with longer sequence length, then PyTorch is forced to\nrelease intermediate buffers from previous iteration and to re-allocate new\nbuffers. This process is time consuming and causes fragmentation in the\ncaching allocator which may result in out-of-memory errors.\n\nA typical solution is to implement pre-allocation. It consists of the\nfollowing steps:\n\n#. generate a (usually random) batch of inputs with maximum sequence length\n (either corresponding to max length in the training dataset or to some\n predefined threshold)\n#. execute a forward and a backward pass with the generated batch, do not\n execute an optimizer or a learning rate scheduler, this step pre-allocates\n buffers of maximum size, which can be reused in subsequent\n training iterations\n#. zero out gradients\n#. proceed to regular training\n\n\n",
"_____no_output_____"
],
[
"Distributed optimizations\n-------------------------\n\n",
"_____no_output_____"
],
[
"Use efficient data-parallel backend\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\nPyTorch has two ways to implement data-parallel training:\n\n* `torch.nn.DataParallel <https://pytorch.org/docs/stable/generated/torch.nn.DataParallel.html#torch.nn.DataParallel>`_\n* `torch.nn.parallel.DistributedDataParallel <https://pytorch.org/docs/stable/generated/torch.nn.parallel.DistributedDataParallel.html#torch.nn.parallel.DistributedDataParallel>`_\n\n``DistributedDataParallel`` offers much better performance and scaling to\nmultiple-GPUs. For more information refer to the\n`relevant section of CUDA Best Practices <https://pytorch.org/docs/stable/notes/cuda.html#use-nn-parallel-distributeddataparallel-instead-of-multiprocessing-or-nn-dataparallel>`_\nfrom PyTorch documentation.\n\n",
"_____no_output_____"
],
[
"Skip unnecessary all-reduce if training with DistributedDataParallel and gradient accumulation\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\nBy default\n`torch.nn.parallel.DistributedDataParallel <https://pytorch.org/docs/stable/generated/torch.nn.parallel.DistributedDataParallel.html#torch.nn.parallel.DistributedDataParallel>`_\nexecutes gradient all-reduce after every backward pass to compute the average\ngradient over all workers participating in the training. If training uses\ngradient accumulation over N steps, then all-reduce is not necessary after\nevery training step, it's only required to perform all-reduce after the last\ncall to backward, just before the execution of the optimizer.\n\n``DistributedDataParallel`` provides\n`no_sync() <https://pytorch.org/docs/stable/generated/torch.nn.parallel.DistributedDataParallel.html#torch.nn.parallel.DistributedDataParallel.no_sync>`_\ncontext manager which disables gradient all-reduce for particular iteration.\n``no_sync()`` should be applied to first ``N-1`` iterations of gradient\naccumulation, the last iteration should follow the default execution and\nperform the required gradient all-reduce.\n\n",
"_____no_output_____"
],
[
"Match the order of layers in constructors and during the execution if using DistributedDataParallel(find_unused_parameters=True)\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n`torch.nn.parallel.DistributedDataParallel <https://pytorch.org/docs/stable/generated/torch.nn.parallel.DistributedDataParallel.html#torch.nn.parallel.DistributedDataParallel>`_\nwith ``find_unused_parameters=True`` uses the order of layers and parameters\nfrom model constructors to build buckets for ``DistributedDataParallel``\ngradient all-reduce. ``DistributedDataParallel`` overlaps all-reduce with the\nbackward pass. All-reduce for a particular bucket is asynchronously triggered\nonly when all gradients for parameters in a given bucket are available.\n\nTo maximize the amount of overlap, the order in model constructors should\nroughly match the order during the execution. If the order doesn't match, then\nall-reduce for the entire bucket waits for the gradient which is the last to\narrive, this may reduce the overlap between backward pass and all-reduce,\nall-reduce may end up being exposed, which slows down the training.\n\n``DistributedDataParallel`` with ``find_unused_parameters=False`` (which is\nthe default setting) relies on automatic bucket formation based on order of\noperations encountered during the backward pass. With\n``find_unused_parameters=False`` it's not necessary to reorder layers or\nparameters to achieve optimal performance.\n\n",
"_____no_output_____"
],
[
"Load-balance workload in a distributed setting\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\nLoad imbalance typically may happen for models processing sequential data\n(speech recognition, translation, language models etc.). If one device\nreceives a batch of data with sequence length longer than sequence lengths for\nthe remaining devices, then all devices wait for the worker which finishes\nlast. Backward pass functions as an implicit synchronization point in a\ndistributed setting with\n`DistributedDataParallel <https://pytorch.org/docs/stable/generated/torch.nn.parallel.DistributedDataParallel.html#torch.nn.parallel.DistributedDataParallel>`_\nbackend.\n\nThere are multiple ways to solve the load balancing problem. The core idea is\nto distribute workload over all workers as uniformly as possible within each\nglobal batch. For example Transformer solves imbalance by forming batches with\napproximately constant number of tokens (and variable number of sequences in a\nbatch), other models solve imbalance by bucketing samples with similar\nsequence length or even by sorting dataset by sequence length.\n\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0011c7e441975c787f600a47e68064de9b44214 | 1,388 | ipynb | Jupyter Notebook | Ref/19.08.2800_mnist_infer_w_80_npart_random/tile.ipynb | danhtaihoang/e-machine | 9ff075ce1e476b8136da291b05abb34c71a4df9d | [
"MIT"
] | null | null | null | Ref/19.08.2800_mnist_infer_w_80_npart_random/tile.ipynb | danhtaihoang/e-machine | 9ff075ce1e476b8136da291b05abb34c71a4df9d | [
"MIT"
] | null | null | null | Ref/19.08.2800_mnist_infer_w_80_npart_random/tile.ipynb | danhtaihoang/e-machine | 9ff075ce1e476b8136da291b05abb34c71a4df9d | [
"MIT"
] | null | null | null | 19.013699 | 36 | 0.399856 | [
[
[
"import numpy as np",
"_____no_output_____"
],
[
"a = [1,2,3,5,7]\nb = np.tile(a,(10,1))\nb",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d0012a6d8953aecb8f770c446f2f90271578226b | 5,687 | ipynb | Jupyter Notebook | leetcode/78_subsets.ipynb | Kaushalya/algo_journal | bcea8afda0dc86b36452378e3bcff9b0f57d6856 | [
"Apache-2.0"
] | null | null | null | leetcode/78_subsets.ipynb | Kaushalya/algo_journal | bcea8afda0dc86b36452378e3bcff9b0f57d6856 | [
"Apache-2.0"
] | null | null | null | leetcode/78_subsets.ipynb | Kaushalya/algo_journal | bcea8afda0dc86b36452378e3bcff9b0f57d6856 | [
"Apache-2.0"
] | null | null | null | 25.733032 | 274 | 0.466151 | [
[
[
"# 78. Subsets\n\n__Difficulty__: Medium\n[Link](https://leetcode.com/problems/subsets/)\n\nGiven an integer array `nums` of unique elements, return all possible subsets (the power set).\n\nThe solution set must not contain duplicate subsets. Return the solution in any order.\n\n__Example 1__:\n\nInput: `nums = [1,2,3]`\nOutput: `[[],[1],[2],[1,2],[3],[1,3],[2,3],[1,2,3]]`\n",
"_____no_output_____"
]
],
[
[
"from typing import List",
"_____no_output_____"
]
],
[
[
"## DFS Approach",
"_____no_output_____"
]
],
[
[
"class SolutionDFS:\n def dfs(self, res, nums):\n if len(nums)==0:\n return [res]\n ans = []\n\n for i, num in enumerate(nums):\n # print(res+[num])\n ans.extend(self.dfs(res+[num], nums[i+1:]))\n ans.append(res)\n # print(ans)\n return ans\n \n def subsets(self, nums: List[int]) -> List[List[int]]:\n return self.dfs([], nums)",
"_____no_output_____"
]
],
[
[
"## Using a bit-mask to indicate selected items from the list of numbers",
"_____no_output_____"
]
],
[
[
"class SolutionMask:\n def subsets(self, nums: List[int]) -> List[List[int]]:\n combs = []\n n = len(nums)\n \n for mask in range(0, 2**n):\n i = 0\n rem = mask\n current_set = []\n while rem:\n if rem%2:\n current_set.append(nums[i])\n rem = rem//2\n i += 1\n combs.append(current_set)\n \n return combs",
"_____no_output_____"
]
],
[
[
"A cleaner and efficient implementation of using bit-mask.",
"_____no_output_____"
]
],
[
[
"class SolutionMask2:\n \n def subsets(self, nums: List[int]) -> List[List[int]]:\n res = []\n n = len(nums)\n nth_bit = 1<<n\n \n for i in range(2**n):\n # To create a bit-mask with length n\n bit_mask = bin(i | nth_bit)[3:]\n res.append([nums[j] for j in range(n) if bit_mask[j]=='1'])\n \n return res",
"_____no_output_____"
]
],
[
[
"## Test cases",
"_____no_output_____"
]
],
[
[
"# Example 1\nnums1 = [1,2,3]\nres1 = [[],[1],[2],[1,2],[3],[1,3],[2,3],[1,2,3]]\n\n# Example 2\nnums2 = [0]\nres2 = [[],[0]]\n\n# Example 3\nnums3 = [0, -2, 5, -7, 9]\nres3 = [[0,-2,5,-7,9],[0,-2,5,-7],[0,-2,5,9],[0,-2,5],[0,-2,-7,9],[0,-2,-7],[0,-2,9],[0,-2],[0,5,-7,9],[0,5,-7],[0,5,9],[0,5],[0,-7,9],[0,-7],[0,9],[0],[-2,5,-7,9],[-2,5,-7],[-2,5,9],[-2,5],[-2,-7,9],[-2,-7],[-2,9],[-2],[5,-7,9],[5,-7],[5,9],[5],[-7,9],[-7],[9],[]]\n\ndef test_function(inp, result):\n assert len(inp)==len(result)\n inp_set = [set(x) for x in inp]\n res_set = [set(x) for x in result]\n\n for i in inp_set:\n assert i in res_set",
"_____no_output_____"
],
[
"# Test DFS method\ndfs_sol = SolutionDFS()\ntest_function(dfs_sol.subsets(nums1), res1)\ntest_function(dfs_sol.subsets(nums2), res2)\ntest_function(dfs_sol.subsets(nums3), res3)",
"_____no_output_____"
],
[
"# Test bit-mask method\nmask_sol = SolutionMask()\ntest_function(mask_sol.subsets(nums1), res1)\ntest_function(mask_sol.subsets(nums2), res2)\ntest_function(mask_sol.subsets(nums3), res3)",
"_____no_output_____"
],
[
"# Test bit-mask method\nmask_sol = SolutionMask2()\ntest_function(mask_sol.subsets(nums1), res1)\ntest_function(mask_sol.subsets(nums2), res2)\ntest_function(mask_sol.subsets(nums3), res3)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0013998bea2ba9edc19dcb49255cb35fa4a9b3b | 5,263 | ipynb | Jupyter Notebook | Matrix Factorization_PySpark/Matrix Factorization Recommendation_PySpark_solution.ipynb | abhisngh/Data-Science | c7fa9e4d81c427382fb9a9d3b97912ef2b21f3ae | [
"MIT"
] | 1 | 2020-05-29T20:07:49.000Z | 2020-05-29T20:07:49.000Z | Matrix Factorization_PySpark/Matrix Factorization Recommendation_PySpark_solution.ipynb | abhisngh/Data-Science | c7fa9e4d81c427382fb9a9d3b97912ef2b21f3ae | [
"MIT"
] | null | null | null | Matrix Factorization_PySpark/Matrix Factorization Recommendation_PySpark_solution.ipynb | abhisngh/Data-Science | c7fa9e4d81c427382fb9a9d3b97912ef2b21f3ae | [
"MIT"
] | null | null | null | 21.569672 | 212 | 0.552157 | [
[
[
"**Matrix factorization** is a class of collaborative filtering algorithms used in recommender systems. **Matrix factorization** approximates a given rating matrix as a product of two lower-rank matrices.\nIt decomposes a rating matrix R(nxm) into a product of two matrices W(nxd) and U(mxd).\n\n\\begin{equation*}\n\\mathbf{R}_{n \\times m} \\approx \\mathbf{\\hat{R}} = \n\\mathbf{V}_{n \\times k} \\times \\mathbf{V}_{m \\times k}^T\n\\end{equation*}",
"_____no_output_____"
]
],
[
[
"#install pyspark\n!pip install pyspark ",
"_____no_output_____"
]
],
[
[
"# Importing the necessary libraries",
"_____no_output_____"
]
],
[
[
"#Import the necessary libraries\nfrom pyspark import SparkContext, SQLContext # required for dealing with dataframes\nimport numpy as np\nfrom pyspark.ml.recommendation import ALS # for Matrix Factorization using ALS ",
"_____no_output_____"
],
[
"# instantiating spark context and SQL context\n",
"_____no_output_____"
]
],
[
[
"#### Step 1. Loading the data into a PySpark dataframe",
"_____no_output_____"
]
],
[
[
"#Read the dataset into a dataframe\njester_ratings_df = sqlContext.read.csv(\"/kaggle/input/jester-17m-jokes-ratings-dataset/jester_ratings.csv\",header = True, inferSchema = True)",
"_____no_output_____"
],
[
"#show the ratings\njester_ratings_df.show(5)",
"_____no_output_____"
],
[
"#Print the total number of ratings, unique users and unique jokes.\n",
"_____no_output_____"
]
],
[
[
"#### Step 2. Splitting into train and test part",
"_____no_output_____"
]
],
[
[
"#Split the dataset using randomSplit in a 90:10 ratio\n",
"_____no_output_____"
],
[
"#Print the training data size and the test data size\n",
"_____no_output_____"
],
[
"#Show the train set\nX_train.show(5)",
"_____no_output_____"
],
[
"#Show the test set\n",
"_____no_output_____"
]
],
[
[
"#### Step 3. Fitting an ALS model",
"_____no_output_____"
]
],
[
[
"#Fit an ALS model with rank=5, maxIter=10 and Seed=0\n",
"_____no_output_____"
],
[
"# displaying the latent features for five users\n ",
"_____no_output_____"
]
],
[
[
"#### Step 4. Making predictions",
"_____no_output_____"
]
],
[
[
"# Pass userId and jokeId from test dataset as an argument \n",
"_____no_output_____"
],
[
"# Join X_test and prediction dataframe and also drop the records for which no predictions are made\n",
"_____no_output_____"
]
],
[
[
"#### Step 5. Evaluating the model",
"_____no_output_____"
]
],
[
[
"# Convert the columns into numpy arrays for direct and easy calculations \n#Also print the RMSE",
"_____no_output_____"
]
],
[
[
"#### Step 6. Recommending jokes",
"_____no_output_____"
]
],
[
[
"# Recommend top 3 jokes for all the users with highest predicted rating \n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0014f81a811c43d11fb4bd3fe7dfee63df9e993 | 43,495 | ipynb | Jupyter Notebook | docs/nb/simplemixing_class.ipynb | IceCubeOpenSource/USSR | d96158cb835245c40e5fc57239c6038c87b3ac01 | [
"BSD-3-Clause"
] | 2 | 2019-05-03T21:09:16.000Z | 2019-11-20T18:40:52.000Z | docs/nb/simplemixing_class.ipynb | IceCubeOpenSource/USSR | d96158cb835245c40e5fc57239c6038c87b3ac01 | [
"BSD-3-Clause"
] | 27 | 2019-03-19T16:02:46.000Z | 2021-07-07T19:36:57.000Z | docs/nb/simplemixing_class.ipynb | IceCubeOpenSource/USSR | d96158cb835245c40e5fc57239c6038c87b3ac01 | [
"BSD-3-Clause"
] | 2 | 2019-03-12T22:36:46.000Z | 2019-05-14T14:14:06.000Z | 231.356383 | 38,796 | 0.912519 | [
[
[
"# Simple Flavor Mixing\n\nIllustrate very basic neutrino flavor mixing in supernova neutrinos using the `SimpleMixing` class in ASTERIA.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\n\nimport astropy.units as u\n\nfrom asteria import config, source\nfrom asteria.neutrino import Flavor\nfrom asteria.oscillation import SimpleMixing\n\nmpl.rc('font', size=16)",
"_____no_output_____"
]
],
[
[
"## Load CCSN Neutrino Model\n\nLoad a neutrino luminosity model (see YAML documentation).",
"_____no_output_____"
]
],
[
[
"conf = config.load_config('../../data/config/test.yaml')\nccsn = source.initialize(conf)",
"_____no_output_____"
]
],
[
[
"## Basic Mixing\n\nSet up the mixing class, which only depends on $\\theta_{12}$.\n\nSee [nu-fit.org](http://www.nu-fit.org/) for current values of the PMNS mixing angles.",
"_____no_output_____"
]
],
[
[
"# Use theta_12 in degrees.\n# To do: explicitly use astropy units for input.\nmix = SimpleMixing(33.8)",
"_____no_output_____"
]
],
[
[
"## Mix the Flavors\n\nApply the mixing and plot the resulting flux curves for the unmixed case and assuming the normal and inverted neutrino mass hierarchies.",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(1, 3, figsize=(13,3.5), sharex=True, sharey=True)\n\nax1, ax2, ax3 = axes\nt = np.linspace(-0.1, 1, 201) * u.s\n\n# UNMIXED\nfor ls, flavor in zip([\"-\", \"--\", \"-.\", \":\"], Flavor):\n flux = ccsn.get_flux(t, flavor)\n \n ax1.plot(t, flux, ls, lw=2, label=flavor.to_tex(), alpha=0.7)\n ax1.set_title(\"Unmixed\")\n# plt.yscale('log')\n# plt.ylim(3e51, 5e53)\nax1.set(xlabel='time - $t_{bounce}$ [s]',\n ylabel='flux')\nax1.legend()\n\n# NORMAL MIXING\nnu_list1 = []\ni = 0\nfor flavor in Flavor:\n flux = ccsn.get_flux(t, flavor)\n nu_list1.append(flux)\n \nnu_new1 = mix.normal_mixing(nu_list1)\n\nfor ls, i, flavor in zip([\"-\", \"--\", \"-.\", \":\"], range(len(nu_new1)), Flavor):\n new_flux1 = nu_new1[i]\n ax2.plot(t, new_flux1, ls, lw=2, alpha=0.7, label=flavor.to_tex())\n ax2.set_title(label=\"Normal Mixing\")\n\nax2.set(xlabel='time - $t_{bounce}$ [s]',\n ylabel='flux')\n\nax2.legend()\n\n# INVERTED MIXING\nnu_list2 = []\ni = 0\nfor flavor in Flavor:\n flux = ccsn.get_flux(t, flavor)\n nu_list2.append(flux)\n \nnu_new2 = mix.inverted_mixing(nu_list1)\n\nfor ls, i, flavor in zip([\"-\", \"--\", \"-.\", \":\"], range(len(nu_new2)), Flavor):\n new_flux2 = nu_new2[i]\n ax3.plot(t, new_flux2, ls, lw=2, alpha=0.7, label=flavor.to_tex())\n ax3.set_title(label=\"Inverted Mixing\")\n\nax3.set(xlabel='time - $t_{bounce}$ [s]',\n ylabel='flux')\nax3.legend()\n\n\nfig.tight_layout();",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0015502b1283645fd893304f6d5181d7499e084 | 8,525 | ipynb | Jupyter Notebook | NotebookExamples/csharp/Samples/HousingML.ipynb | nelson-wu/try | f133e7d20b1cf1384b313f9ddee9fc70bc6aa055 | [
"MIT"
] | 2 | 2019-12-06T07:55:50.000Z | 2019-12-06T07:57:57.000Z | NotebookExamples/csharp/Samples/HousingML.ipynb | https-microsoft-com-powershell/try | 4b94491d734c9f0928ab82663ca642d07e35f10f | [
"MIT"
] | null | null | null | NotebookExamples/csharp/Samples/HousingML.ipynb | https-microsoft-com-powershell/try | 4b94491d734c9f0928ab82663ca642d07e35f10f | [
"MIT"
] | null | null | null | 25.600601 | 134 | 0.494194 | [
[
[
"#r \"nuget:Microsoft.ML,1.4.0\"\n#r \"nuget:Microsoft.ML.AutoML,0.16.0\"\n#r \"nuget:Microsoft.Data.Analysis,0.1.0\"",
"_____no_output_____"
],
[
"using Microsoft.Data.Analysis;\nusing XPlot.Plotly;",
"_____no_output_____"
],
[
"using Microsoft.AspNetCore.Html;\nFormatter<DataFrame>.Register((df, writer) =>\n{\n var headers = new List<IHtmlContent>();\n headers.Add(th(i(\"index\")));\n headers.AddRange(df.Columns.Select(c => (IHtmlContent) th(c.Name)));\n var rows = new List<List<IHtmlContent>>();\n var take = 20;\n for (var i = 0; i < Math.Min(take, df.RowCount); i++)\n {\n var cells = new List<IHtmlContent>();\n cells.Add(td(i));\n foreach (var obj in df[i])\n {\n cells.Add(td(obj));\n }\n rows.Add(cells);\n }\n \n var t = table(\n thead(\n headers),\n tbody(\n rows.Select(\n r => tr(r))));\n \n writer.Write(t);\n}, \"text/html\");",
"_____no_output_____"
],
[
"using System.IO;\nusing System.Net.Http;\nstring housingPath = \"housing.csv\";\nif (!File.Exists(housingPath))\n{\n var contents = new HttpClient()\n .GetStringAsync(\"https://raw.githubusercontent.com/ageron/handson-ml2/master/datasets/housing/housing.csv\").Result;\n \n File.WriteAllText(\"housing.csv\", contents);\n}",
"_____no_output_____"
],
[
"var housingData = DataFrame.LoadCsv(housingPath);\nhousingData",
"_____no_output_____"
],
[
"housingData.Description()",
"_____no_output_____"
],
[
"Chart.Plot(\n new Graph.Histogram()\n {\n x = housingData[\"median_house_value\"],\n nbinsx = 20\n }\n)",
"_____no_output_____"
],
[
"var chart = Chart.Plot(\n new Graph.Scattergl()\n {\n x = housingData[\"longitude\"],\n y = housingData[\"latitude\"],\n mode = \"markers\",\n marker = new Graph.Marker()\n {\n color = housingData[\"median_house_value\"],\n colorscale = \"Jet\"\n }\n }\n);\n\nchart.Width = 600;\nchart.Height = 600;\ndisplay(chart);",
"_____no_output_____"
],
[
"static T[] Shuffle<T>(T[] array)\n{\n Random rand = new Random();\n for (int i = 0; i < array.Length; i++)\n {\n int r = i + rand.Next(array.Length - i);\n T temp = array[r];\n array[r] = array[i];\n array[i] = temp;\n }\n return array;\n}\n\nint[] randomIndices = Shuffle(Enumerable.Range(0, (int)housingData.RowCount).ToArray());\nint testSize = (int)(housingData.RowCount * .1);\nint[] trainRows = randomIndices[testSize..];\nint[] testRows = randomIndices[..testSize];\n\nDataFrame housing_train = housingData[trainRows];\nDataFrame housing_test = housingData[testRows];\n\ndisplay(housing_train.RowCount);\ndisplay(housing_test.RowCount);",
"_____no_output_____"
],
[
"using Microsoft.ML;\nusing Microsoft.ML.Data;\nusing Microsoft.ML.AutoML;",
"_____no_output_____"
],
[
"%%time\n\nvar mlContext = new MLContext();\n\nvar experiment = mlContext.Auto().CreateRegressionExperiment(maxExperimentTimeInSeconds: 15);\nvar result = experiment.Execute(housing_train, labelColumnName:\"median_house_value\");",
"_____no_output_____"
],
[
"var scatters = result.RunDetails.Where(d => d.ValidationMetrics != null).GroupBy( \n r => r.TrainerName,\n (name, details) => new Graph.Scattergl()\n {\n name = name,\n x = details.Select(r => r.RuntimeInSeconds),\n y = details.Select(r => r.ValidationMetrics.MeanAbsoluteError),\n mode = \"markers\",\n marker = new Graph.Marker() { size = 12 }\n });\n\nvar chart = Chart.Plot(scatters);\nchart.WithXTitle(\"Training Time\");\nchart.WithYTitle(\"Error\");\ndisplay(chart);\n\nConsole.WriteLine($\"Best Trainer:{result.BestRun.TrainerName}\");",
"_____no_output_____"
],
[
"var testResults = result.BestRun.Model.Transform(housing_test);\n\nvar trueValues = testResults.GetColumn<float>(\"median_house_value\");\nvar predictedValues = testResults.GetColumn<float>(\"Score\");\n\nvar predictedVsTrue = new Graph.Scattergl()\n{\n x = trueValues,\n y = predictedValues,\n mode = \"markers\",\n};\n\nvar maximumValue = Math.Max(trueValues.Max(), predictedValues.Max());\n\nvar perfectLine = new Graph.Scattergl()\n{\n x = new[] {0, maximumValue},\n y = new[] {0, maximumValue},\n mode = \"lines\",\n};\n\nvar chart = Chart.Plot(new[] {predictedVsTrue, perfectLine });\nchart.WithXTitle(\"True Values\");\nchart.WithYTitle(\"Predicted Values\");\nchart.WithLegend(false);\nchart.Width = 600;\nchart.Height = 600;\ndisplay(chart);",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0015f52f5ed4adc43789892f3c2a6aa972b6023 | 26,788 | ipynb | Jupyter Notebook | Pertemuan 8/Imputasi.ipynb | grudasakti/metdat-science | 3f3aa7e5e9390c27a8b2eab66d754e251a8ff342 | [
"MIT"
] | null | null | null | Pertemuan 8/Imputasi.ipynb | grudasakti/metdat-science | 3f3aa7e5e9390c27a8b2eab66d754e251a8ff342 | [
"MIT"
] | null | null | null | Pertemuan 8/Imputasi.ipynb | grudasakti/metdat-science | 3f3aa7e5e9390c27a8b2eab66d754e251a8ff342 | [
"MIT"
] | null | null | null | 25.391469 | 171 | 0.359788 | [
[
[
"# Imputasi",
"_____no_output_____"
],
[
"Imputasi adalah mengganti nilai/data yang hilang (missing value; NaN; blank) dengan nilai pengganti",
"_____no_output_____"
],
[
"### Mean",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nkolom = {'col1':[2, 9, 19],\n 'col2':[5, np.nan, 17],\n 'col3':[3, 9, np.nan],\n 'col4':[6, 0, 9],\n 'col5':[np.nan, 7, np.nan]}\n\ndata = pd.DataFrame(kolom)",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"data.fillna(data.mean())",
"_____no_output_____"
]
],
[
[
"### Arbitrary (Nilai Suka-Suka)",
"_____no_output_____"
]
],
[
[
"umur = {'umur' : [29, 43, np.nan, 25, 34, np.nan, 50]}\n\ndata = pd.DataFrame(umur)\ndata",
"_____no_output_____"
],
[
"data.fillna(99)",
"_____no_output_____"
]
],
[
[
"### End of Tail",
"_____no_output_____"
]
],
[
[
"umur = {'umur' : [29, 43, np.nan, 25, 34, np.nan, 50]}\n\ndata = pd.DataFrame(umur)\ndata",
"_____no_output_____"
],
[
"#install library feature-engine\npip install feature-engine",
"Collecting feature-engineNote: you may need to restart the kernel to use updated packages.\n Downloading feature_engine-1.2.0-py2.py3-none-any.whl (205 kB)\nRequirement already satisfied: numpy>=1.18.2 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from feature-engine) (1.20.1)\n\nRequirement already satisfied: scipy>=1.4.1 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from feature-engine) (1.6.2)\nRequirement already satisfied: pandas>=1.0.3 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from feature-engine) (1.2.4)\nRequirement already satisfied: scikit-learn>=0.22.2 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from feature-engine) (0.24.1)\nRequirement already satisfied: statsmodels>=0.11.1 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from feature-engine) (0.12.2)\nRequirement already satisfied: pytz>=2017.3 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from pandas>=1.0.3->feature-engine) (2021.1)\nRequirement already satisfied: python-dateutil>=2.7.3 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from pandas>=1.0.3->feature-engine) (2.8.1)\nRequirement already satisfied: six>=1.5 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from python-dateutil>=2.7.3->pandas>=1.0.3->feature-engine) (1.15.0)\nRequirement already satisfied: joblib>=0.11 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from scikit-learn>=0.22.2->feature-engine) (1.0.1)\nRequirement already satisfied: threadpoolctl>=2.0.0 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from scikit-learn>=0.22.2->feature-engine) (2.1.0)\nRequirement already satisfied: patsy>=0.5 in c:\\users\\gruda\\anaconda3\\lib\\site-packages (from statsmodels>=0.11.1->feature-engine) (0.5.1)\nInstalling collected packages: feature-engine\nSuccessfully installed feature-engine-1.2.0\n"
],
[
"#import EndTailImputer\nfrom feature_engine.imputation import EndTailImputer\n\n#buat Imputer\nimputer = EndTailImputer(imputation_method='gaussian', tail='right')\n\n#fit-kan imputer ke set\nimputer.fit(data)\n\n#ubah data\ntest_data = imputer.transform(data)\n\n#tampil data\ntest_data",
"_____no_output_____"
]
],
[
[
"## Data Kategorikal",
"_____no_output_____"
],
[
"### Modus",
"_____no_output_____"
]
],
[
[
"from sklearn.impute import SimpleImputer\nmobil = {'mobil':['Ford', 'Ford', 'Toyota', 'Honda', np.nan, 'Toyota', 'Honda', 'Toyota', np.nan, np.nan]}\n\ndata = pd.DataFrame(mobil)",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"imp = SimpleImputer(strategy='most_frequent')",
"_____no_output_____"
],
[
"imp.fit_transform(data)",
"_____no_output_____"
]
],
[
[
"### Random Sample",
"_____no_output_____"
]
],
[
[
"#import Random Sample\nfrom feature_engine.imputation import RandomSampleImputer\n\n#buat data missing value\ndata = {'Jenis Kelamin' : ['Laki-laki', 'Perempuan', 'Laki-laki', np.nan, 'Laki-laki', 'Perempuan', 'Perempuan', np.nan, 'Laki-laki', np.nan],\n 'Umur' : [29, np.nan, 32, 43, 50, 22, np.nan, 52, np.nan, 17]}\n\ndf = pd.DataFrame(data)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"#membuat imputer\nimputer = RandomSampleImputer(random_state=29)\n\n#fit-kan\nimputer.fit(df)\n\n#ubah data\ntesting_df = imputer.transform(df)",
"_____no_output_____"
],
[
"testing_df",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0017a1caaf8a05aa05585355f7f2498bbd24229 | 39,679 | ipynb | Jupyter Notebook | chap11.ipynb | dev-strender/python-machine-learning-with-scikit-learn | acac8032e831b72ed6a84bd57d03a958d4bb09b0 | [
"MIT"
] | 1 | 2019-10-25T03:26:57.000Z | 2019-10-25T03:26:57.000Z | chap11.ipynb | dev-strender/python-machine-learning-with-scikit-learn | acac8032e831b72ed6a84bd57d03a958d4bb09b0 | [
"MIT"
] | null | null | null | chap11.ipynb | dev-strender/python-machine-learning-with-scikit-learn | acac8032e831b72ed6a84bd57d03a958d4bb09b0 | [
"MIT"
] | null | null | null | 30.105463 | 1,351 | 0.538673 | [
[
[
"# Chapter 8 - Applying Machine Learning To Sentiment Analysis",
"_____no_output_____"
],
[
"### Overview",
"_____no_output_____"
],
[
"- [Obtaining the IMDb movie review dataset](#Obtaining-the-IMDb-movie-review-dataset)\n- [Introducing the bag-of-words model](#Introducing-the-bag-of-words-model)\n - [Transforming words into feature vectors](#Transforming-words-into-feature-vectors)\n - [Assessing word relevancy via term frequency-inverse document frequency](#Assessing-word-relevancy-via-term-frequency-inverse-document-frequency)\n - [Cleaning text data](#Cleaning-text-data)\n - [Processing documents into tokens](#Processing-documents-into-tokens)\n- [Training a logistic regression model for document classification](#Training-a-logistic-regression-model-for-document-classification)\n- [Working with bigger data – online algorithms and out-of-core learning](#Working-with-bigger-data-–-online-algorithms-and-out-of-core-learning)\n- [Summary](#Summary)",
"_____no_output_____"
],
[
"NLP: Natural Language Processing ",
"_____no_output_____"
],
[
"#### Sentiment Analysis (Opinion Mining)\nAnalyzes the polarity of documents\n- Expressed opinions or emotions of the authors with regard to a particular topic",
"_____no_output_____"
],
[
"# Obtaining the IMDb movie review dataset",
"_____no_output_____"
],
[
"- IMDb: the Internet Movie Database\n- IMDb dataset\n - A. L. Maas, R. E. Daly, P. T. Pham, D. Huang, A. Y. Ng, and C. Potts. Learning Word Vectors for Sentiment Analysis. In the proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies, pages 142–150, Portland, Oregon, USA, June 2011. Association for Computational Linguistics\n- 50,000 movie reviews labeled either *positive* or *negative*",
"_____no_output_____"
],
[
"The IMDB movie review set can be downloaded from [http://ai.stanford.edu/~amaas/data/sentiment/](http://ai.stanford.edu/~amaas/data/sentiment/).\nAfter downloading the dataset, decompress the files.\n\n`aclImdb_v1.tar.gz`\n",
"_____no_output_____"
]
],
[
[
"import pyprind\nimport pandas as pd\nimport os\n\n# change the `basepath` to the directory of the\n# unzipped movie dataset\n\nbasepath = '/Users/sklee/datasets/aclImdb/'\n\nlabels = {'pos': 1, 'neg': 0}\npbar = pyprind.ProgBar(50000)\ndf = pd.DataFrame()\nfor s in ('test', 'train'):\n for l in ('pos', 'neg'):\n path = os.path.join(basepath, s, l)\n for file in os.listdir(path):\n with open(os.path.join(path, file), 'r', encoding='utf-8') as infile:\n txt = infile.read()\n df = df.append([[txt, labels[l]]], ignore_index=True)\n pbar.update()\ndf.columns = ['review', 'sentiment']",
"0% [##############################] 100% | ETA: 00:00:00\nTotal time elapsed: 00:01:58\n"
],
[
"df.head(5)",
"_____no_output_____"
]
],
[
[
"Shuffling the DataFrame:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nnp.random.seed(0)\ndf = df.reindex(np.random.permutation(df.index))",
"_____no_output_____"
],
[
"df.head(5)",
"_____no_output_____"
],
[
"df.to_csv('./movie_data.csv', index=False)",
"_____no_output_____"
]
],
[
[
"<br>\n<br>",
"_____no_output_____"
],
[
"# Introducing the bag-of-words model",
"_____no_output_____"
],
[
"- **Vocabulary** : the collection of unique tokens (e.g. words) from the entire set of documents\n- Construct a feature vector from each document\n - Vector length = length of the vocabulary\n - Contains the counts of how often each token occurs in the particular document\n - Sparse vectors",
"_____no_output_____"
],
[
"## Transforming documents into feature vectors",
"_____no_output_____"
],
[
"By calling the fit_transform method on CountVectorizer, we just constructed the vocabulary of the bag-of-words model and transformed the following three sentences into sparse feature vectors:\n1. The sun is shining\n2. The weather is sweet\n3. The sun is shining, the weather is sweet, and one and one is two\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.feature_extraction.text import CountVectorizer\n\ncount = CountVectorizer()\ndocs = np.array([\n 'The sun is shining',\n 'The weather is sweet',\n 'The sun is shining, the weather is sweet, and one and one is two'])\nbag = count.fit_transform(docs)",
"_____no_output_____"
]
],
[
[
"Now let us print the contents of the vocabulary to get a better understanding of the underlying concepts:",
"_____no_output_____"
]
],
[
[
"print(count.vocabulary_)",
"{'the': 6, 'sun': 4, 'is': 1, 'shining': 3, 'weather': 8, 'sweet': 5, 'and': 0, 'one': 2, 'two': 7}\n"
]
],
[
[
"As we can see from executing the preceding command, the vocabulary is stored in a Python dictionary, which maps the unique words that are mapped to integer indices. Next let us print the feature vectors that we just created:",
"_____no_output_____"
],
[
"Each index position in the feature vectors shown here corresponds to the integer values that are stored as dictionary items in the CountVectorizer vocabulary. For example, the rst feature at index position 0 resembles the count of the word and, which only occurs in the last document, and the word is at index position 1 (the 2nd feature in the document vectors) occurs in all three sentences. Those values in the feature vectors are also called the raw term frequencies: *tf (t,d)*—the number of times a term t occurs in a document *d*.",
"_____no_output_____"
]
],
[
[
"print(bag.toarray())",
"[[0 1 0 1 1 0 1 0 0]\n [0 1 0 0 0 1 1 0 1]\n [2 3 2 1 1 1 2 1 1]]\n"
]
],
[
[
"Those count values are called the **raw term frequency td(t,d)**\n - t: term\n - d: document",
"_____no_output_____"
],
[
"The **n-gram** Models\n- 1-gram: \"the\", \"sun\", \"is\", \"shining\"\n- 2-gram: \"the sun\", \"sun is\", \"is shining\"\n - CountVectorizer(ngram_range=(2,2))",
"_____no_output_____"
],
[
"<br>",
"_____no_output_____"
],
[
"## Assessing word relevancy via term frequency-inverse document frequency",
"_____no_output_____"
]
],
[
[
"np.set_printoptions(precision=2)",
"_____no_output_____"
]
],
[
[
"- Frequently occurring words across multiple documents from both classes typically don't contain useful or discriminatory information. \n- ** Term frequency-inverse document frequency (tf-idf)** can be used to downweight those frequently occurring words in the feature vectors.\n\n$$\\text{tf-idf}(t,d)=\\text{tf (t,d)}\\times \\text{idf}(t,d)$$\n\n - **tf(t, d) the term frequency**\n - **idf(t, d) the inverse document frequency**:\n\n$$\\text{idf}(t,d) = \\text{log}\\frac{n_d}{1+\\text{df}(d, t)},$$\n \n - $n_d$ is the total number of documents\n - **df(d, t) document frequency**: the number of documents *d* that contain the term *t*. \n - Note that adding the constant 1 to the denominator is optional and serves the purpose of assigning a non-zero value to terms that occur in all training samples; the log is used to ensure that low document frequencies are not given too much weight.\n\nScikit-learn implements yet another transformer, the `TfidfTransformer`, that takes the raw term frequencies from `CountVectorizer` as input and transforms them into tf-idfs:",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import TfidfTransformer\n\ntfidf = TfidfTransformer(use_idf=True, norm='l2', smooth_idf=True)\nprint(tfidf.fit_transform(count.fit_transform(docs)).toarray())",
"[[ 0. 0.43 0. 0.56 0.56 0. 0.43 0. 0. ]\n [ 0. 0.43 0. 0. 0. 0.56 0.43 0. 0.56]\n [ 0.5 0.45 0.5 0.19 0.19 0.19 0.3 0.25 0.19]]\n"
]
],
[
[
"As we saw in the previous subsection, the word is had the largest term frequency in the 3rd document, being the most frequently occurring word. However, after transforming the same feature vector into tf-idfs, we see that the word is is\nnow associated with a relatively small tf-idf (0.45) in document 3 since it is\nalso contained in documents 1 and 2 and thus is unlikely to contain any useful, discriminatory information.\n",
"_____no_output_____"
],
[
"However, if we'd manually calculated the tf-idfs of the individual terms in our feature vectors, we'd have noticed that the `TfidfTransformer` calculates the tf-idfs slightly differently compared to the standard textbook equations that we de ned earlier. The equations for the idf and tf-idf that were implemented in scikit-learn are:",
"_____no_output_____"
],
[
"$$\\text{idf} (t,d) = log\\frac{1 + n_d}{1 + \\text{df}(d, t)}$$\n\nThe tf-idf equation that was implemented in scikit-learn is as follows:\n\n$$\\text{tf-idf}(t,d) = \\text{tf}(t,d) \\times (\\text{idf}(t,d)+1)$$\n\nWhile it is also more typical to normalize the raw term frequencies before calculating the tf-idfs, the `TfidfTransformer` normalizes the tf-idfs directly.\n\nBy default (`norm='l2'`), scikit-learn's TfidfTransformer applies the L2-normalization, which returns a vector of length 1 by dividing an un-normalized feature vector *v* by its L2-norm:\n\n$$v_{\\text{norm}} = \\frac{v}{||v||_2} = \\frac{v}{\\sqrt{v_{1}^{2} + v_{2}^{2} + \\dots + v_{n}^{2}}} = \\frac{v}{\\big (\\sum_{i=1}^{n} v_{i}^{2}\\big)^\\frac{1}{2}}$$\n\nTo make sure that we understand how TfidfTransformer works, let us walk\nthrough an example and calculate the tf-idf of the word is in the 3rd document.\n\nThe word is has a term frequency of 3 (tf = 3) in document 3, and the document frequency of this term is 3 since the term is occurs in all three documents (df = 3). Thus, we can calculate the idf as follows:\n\n$$\\text{idf}(\"is\", d3) = log \\frac{1+3}{1+3} = 0$$\n\nNow in order to calculate the tf-idf, we simply need to add 1 to the inverse document frequency and multiply it by the term frequency:\n\n$$\\text{tf-idf}(\"is\",d3)= 3 \\times (0+1) = 3$$",
"_____no_output_____"
]
],
[
[
"tf_is = 3\nn_docs = 3\nidf_is = np.log((n_docs+1) / (3+1))\ntfidf_is = tf_is * (idf_is + 1)\nprint('tf-idf of term \"is\" = %.2f' % tfidf_is)",
"tf-idf of term \"is\" = 3.00\n"
]
],
[
[
"If we repeated these calculations for all terms in the 3rd document, we'd obtain the following tf-idf vectors: [3.39, 3.0, 3.39, 1.29, 1.29, 1.29, 2.0 , 1.69, 1.29]. However, we notice that the values in this feature vector are different from the values that we obtained from the TfidfTransformer that we used previously. The nal step that we are missing in this tf-idf calculation is the L2-normalization, which can be applied as follows:",
"_____no_output_____"
],
[
"$$\\text{tfi-df}_{norm} = \\frac{[3.39, 3.0, 3.39, 1.29, 1.29, 1.29, 2.0 , 1.69, 1.29]}{\\sqrt{[3.39^2, 3.0^2, 3.39^2, 1.29^2, 1.29^2, 1.29^2, 2.0^2 , 1.69^2, 1.29^2]}}$$\n\n$$=[0.5, 0.45, 0.5, 0.19, 0.19, 0.19, 0.3, 0.25, 0.19]$$\n\n$$\\Rightarrow \\text{tf-idf}_{norm}(\"is\", d3) = 0.45$$",
"_____no_output_____"
],
[
"As we can see, the results match the results returned by scikit-learn's `TfidfTransformer` (below). Since we now understand how tf-idfs are calculated, let us proceed to the next sections and apply those concepts to the movie review dataset.",
"_____no_output_____"
]
],
[
[
"tfidf = TfidfTransformer(use_idf=True, norm=None, smooth_idf=True)\nraw_tfidf = tfidf.fit_transform(count.fit_transform(docs)).toarray()[-1]\nraw_tfidf ",
"_____no_output_____"
],
[
"l2_tfidf = raw_tfidf / np.sqrt(np.sum(raw_tfidf**2))\nl2_tfidf",
"_____no_output_____"
]
],
[
[
"<br>",
"_____no_output_____"
],
[
"## Cleaning text data",
"_____no_output_____"
],
[
"**Before** we build the bag-of-words model.",
"_____no_output_____"
]
],
[
[
"df.loc[112, 'review'][-1000:]",
"_____no_output_____"
]
],
[
[
"#### Python regular expression library",
"_____no_output_____"
]
],
[
[
"import re\ndef preprocessor(text):\n text = re.sub('<[^>]*>', '', text)\n emoticons = re.findall('(?::|;|=)(?:-)?(?:\\)|\\(|D|P)', text)\n text = re.sub('[\\W]+', ' ', text.lower()) +\\\n ' '.join(emoticons).replace('-', '')\n return text",
"_____no_output_____"
],
[
"preprocessor(df.loc[112, 'review'][-1000:])",
"_____no_output_____"
],
[
"preprocessor(\"</a>This :) is :( a test :-)!\")",
"_____no_output_____"
],
[
"df['review'] = df['review'].apply(preprocessor)",
"_____no_output_____"
]
],
[
[
"<br>",
"_____no_output_____"
],
[
"## Processing documents into tokens",
"_____no_output_____"
],
[
"#### Word Stemming\nTransforming a word into its root form\n\n- Original stemming algorithm: Martin F. Porter. An algorithm for suf x stripping. Program: electronic library and information systems, 14(3):130–137, 1980)\n- Snowball stemmer (Porter2 or \"English\" stemmer)\n- Lancaster stemmer (Paice-Husk stemmer)",
"_____no_output_____"
],
[
"Python NLP toolkit: NLTK (the Natural Language ToolKit)\n - Free online book http://www.nltk.org/book/",
"_____no_output_____"
]
],
[
[
"from nltk.stem.porter import PorterStemmer\n\nporter = PorterStemmer()\n\ndef tokenizer(text):\n return text.split()\n\n\ndef tokenizer_porter(text):\n return [porter.stem(word) for word in text.split()]",
"_____no_output_____"
],
[
"tokenizer('runners like running and thus they run')",
"_____no_output_____"
],
[
"tokenizer_porter('runners like running and thus they run')",
"_____no_output_____"
]
],
[
[
"#### Lemmatization\n- thus -> thu\n- Tries to find canonical forms of words\n- Computationally expensive, little impact on text classification performance",
"_____no_output_____"
],
[
"#### Stop-words Removal\n- Stop-words: extremely common words, e.g., is, and, has, like...\n- Removal is useful when we use raw or normalized tf, rather than tf-idf",
"_____no_output_____"
]
],
[
[
"import nltk\n\nnltk.download('stopwords')",
"[nltk_data] Downloading package stopwords to /Users/sklee/nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
],
[
"from nltk.corpus import stopwords\n\nstop = stopwords.words('english')\n[w for w in tokenizer_porter('a runner likes running and runs a lot')[-10:]\nif w not in stop]",
"_____no_output_____"
],
[
"stop[-10:]",
"_____no_output_____"
]
],
[
[
"<br>\n<br>",
"_____no_output_____"
],
[
"# Training a logistic regression model for document classification",
"_____no_output_____"
]
],
[
[
"X_train = df.loc[:25000, 'review'].values\ny_train = df.loc[:25000, 'sentiment'].values\nX_test = df.loc[25000:, 'review'].values\ny_test = df.loc[25000:, 'sentiment'].values",
"_____no_output_____"
],
[
"from sklearn.pipeline import Pipeline\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.model_selection import GridSearchCV\n\ntfidf = TfidfVectorizer(strip_accents=None,\n lowercase=False,\n preprocessor=None)\n\nparam_grid = [{'vect__ngram_range': [(1, 1)],\n 'vect__stop_words': [stop, None],\n 'vect__tokenizer': [tokenizer, tokenizer_porter],\n 'clf__penalty': ['l1', 'l2'],\n 'clf__C': [1.0, 10.0, 100.0]},\n {'vect__ngram_range': [(1, 1)],\n 'vect__stop_words': [stop, None],\n 'vect__tokenizer': [tokenizer, tokenizer_porter],\n 'vect__use_idf':[False],\n 'vect__norm':[None],\n 'clf__penalty': ['l1', 'l2'],\n 'clf__C': [1.0, 10.0, 100.0]},\n ]\n\nlr_tfidf = Pipeline([('vect', tfidf),\n ('clf', LogisticRegression(random_state=0))])\n\ngs_lr_tfidf = GridSearchCV(lr_tfidf, param_grid,\n scoring='accuracy',\n cv=5,\n verbose=1,\n n_jobs=-1)",
"_____no_output_____"
],
[
"gs_lr_tfidf.fit(X_train, y_train)",
"Fitting 5 folds for each of 48 candidates, totalling 240 fits\n"
],
[
"print('Best parameter set: %s ' % gs_lr_tfidf.best_params_)\nprint('CV Accuracy: %.3f' % gs_lr_tfidf.best_score_)\n# Best parameter set: {'vect__tokenizer': <function tokenizer at 0x11851c6a8>, 'clf__C': 10.0, 'vect__stop_words': None, 'clf__penalty': 'l2', 'vect__ngram_range': (1, 1)} \n# CV Accuracy: 0.897",
"_____no_output_____"
],
[
"clf = gs_lr_tfidf.best_estimator_\nprint('Test Accuracy: %.3f' % clf.score(X_test, y_test))\n# Test Accuracy: 0.899",
"_____no_output_____"
]
],
[
[
"<br>\n<br>",
"_____no_output_____"
],
[
"# Working with bigger data - online algorithms and out-of-core learning",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport re\nfrom nltk.corpus import stopwords\n\ndef tokenizer(text):\n text = re.sub('<[^>]*>', '', text)\n emoticons = re.findall('(?::|;|=)(?:-)?(?:\\)|\\(|D|P)', text.lower())\n text = re.sub('[\\W]+', ' ', text.lower()) +\\\n ' '.join(emoticons).replace('-', '')\n tokenized = [w for w in text.split() if w not in stop]\n return tokenized\n\n\n# reads in and returns one document at a time\ndef stream_docs(path):\n with open(path, 'r', encoding='utf-8') as csv:\n next(csv) # skip header\n for line in csv:\n text, label = line[:-3], int(line[-2])\n yield text, label",
"_____no_output_____"
],
[
"doc_stream = stream_docs(path='./movie_data.csv')",
"_____no_output_____"
],
[
"next(doc_stream)",
"_____no_output_____"
]
],
[
[
"#### Minibatch",
"_____no_output_____"
]
],
[
[
"def get_minibatch(doc_stream, size):\n docs, y = [], []\n try:\n for _ in range(size):\n text, label = next(doc_stream)\n docs.append(text)\n y.append(label)\n except StopIteration:\n return None, None\n return docs, y",
"_____no_output_____"
]
],
[
[
"- We cannot use `CountVectorizer` since it requires holding the complete vocabulary. Likewise, `TfidfVectorizer` needs to keep all feature vectors in memory. \n\n- We can use `HashingVectorizer` instead for online training (32-bit MurmurHash3 algorithm by Austin Appleby (https://sites.google.com/site/murmurhash/)",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import HashingVectorizer\nfrom sklearn.linear_model import SGDClassifier\n\nvect = HashingVectorizer(decode_error='ignore', \n n_features=2**21,\n preprocessor=None, \n tokenizer=tokenizer)\n\nclf = SGDClassifier(loss='log', random_state=1, max_iter=1)\ndoc_stream = stream_docs(path='./movie_data.csv')",
"_____no_output_____"
],
[
"import pyprind\npbar = pyprind.ProgBar(45)\n\nclasses = np.array([0, 1])\nfor _ in range(45):\n X_train, y_train = get_minibatch(doc_stream, size=1000)\n if not X_train:\n break\n X_train = vect.transform(X_train)\n clf.partial_fit(X_train, y_train, classes=classes)\n pbar.update()",
"0% [##############################] 100% | ETA: 00:00:00\nTotal time elapsed: 00:00:23\n"
],
[
"X_test, y_test = get_minibatch(doc_stream, size=5000)\nX_test = vect.transform(X_test)\nprint('Accuracy: %.3f' % clf.score(X_test, y_test))",
"Accuracy: 0.867\n"
],
[
"clf = clf.partial_fit(X_test, y_test)",
"_____no_output_____"
]
],
[
[
"<br>\n<br>",
"_____no_output_____"
],
[
"# Summary",
"_____no_output_____"
],
[
"- **Latent Dirichlet allocation**, a topic model that considers the latent semantics of words (D. M. Blei, A. Y. Ng, and M. I. Jordan. Latent Dirichlet allocation. The Journal of machine Learning research, 3:993–1022, 2003)\n- **word2vec**, an algorithm that Google released in 2013 (T. Mikolov, K. Chen, G. Corrado, and J. Dean. Ef cient Estimation of Word Representations in Vector Space. arXiv preprint arXiv:1301.3781, 2013)\n - https://code.google.com/p/word2vec/.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0018c80b3442519c1eb803e38228645f34732bc | 617,043 | ipynb | Jupyter Notebook | 09_NLP_Evaluation/ClassificationEvaluation.ipynb | satyajitghana/TSAI-DeepNLP-END2.0 | de5faeb8a3d266346e6e62f75b8f64c5514053bb | [
"MIT"
] | 1 | 2021-06-08T14:41:40.000Z | 2021-06-08T14:41:40.000Z | 09_NLP_Evaluation/ClassificationEvaluation.ipynb | satyajitghana/TSAI-DeepNLP-END2.0 | de5faeb8a3d266346e6e62f75b8f64c5514053bb | [
"MIT"
] | null | null | null | 09_NLP_Evaluation/ClassificationEvaluation.ipynb | satyajitghana/TSAI-DeepNLP-END2.0 | de5faeb8a3d266346e6e62f75b8f64c5514053bb | [
"MIT"
] | 8 | 2021-05-12T17:40:25.000Z | 2022-01-20T14:38:43.000Z | 119.396865 | 44,014 | 0.817734 | [
[
[
"<a href=\"https://colab.research.google.com/github/satyajitghana/TSAI-DeepNLP-END2.0/blob/main/09_NLP_Evaluation/ClassificationEvaluation.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"! pip3 install git+https://github.com/extensive-nlp/ttc_nlp --quiet\n! pip3 install torchmetrics --quiet",
" Installing build dependencies ... \u001b[?25l\u001b[?25hdone\n Getting requirements to build wheel ... \u001b[?25l\u001b[?25hdone\n Preparing wheel metadata ... \u001b[?25l\u001b[?25hdone\n\u001b[K |████████████████████████████████| 6.4MB 8.0MB/s \n\u001b[?25h Installing build dependencies ... \u001b[?25l\u001b[?25hdone\n Getting requirements to build wheel ... \u001b[?25l\u001b[?25hdone\n Preparing wheel metadata ... \u001b[?25l\u001b[?25hdone\n\u001b[K |████████████████████████████████| 81kB 10.6MB/s \n\u001b[K |████████████████████████████████| 10.3MB 46.7MB/s \n\u001b[K |████████████████████████████████| 10.8MB 188kB/s \n\u001b[K |████████████████████████████████| 819kB 37.3MB/s \n\u001b[K |████████████████████████████████| 81kB 11.5MB/s \n\u001b[K |████████████████████████████████| 51kB 8.0MB/s \n\u001b[K |████████████████████████████████| 624kB 42.7MB/s \n\u001b[K |████████████████████████████████| 10.1MB 46.9MB/s \n\u001b[K |████████████████████████████████| 460kB 45.3MB/s \n\u001b[K |████████████████████████████████| 645kB 38.8MB/s \n\u001b[K |████████████████████████████████| 112kB 55.0MB/s \n\u001b[K |████████████████████████████████| 235kB 52.7MB/s \n\u001b[K |████████████████████████████████| 122kB 38.2MB/s \n\u001b[K |████████████████████████████████| 10.6MB 40.7MB/s \n\u001b[K |████████████████████████████████| 829kB 38.0MB/s \n\u001b[K |████████████████████████████████| 1.3MB 33.2MB/s \n\u001b[K |████████████████████████████████| 296kB 56.6MB/s \n\u001b[K |████████████████████████████████| 143kB 56.1MB/s \n\u001b[?25h Building wheel for ttctext (PEP 517) ... \u001b[?25l\u001b[?25hdone\n Building wheel for gdown (PEP 517) ... \u001b[?25l\u001b[?25hdone\n Building wheel for antlr4-python3-runtime (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for future (setup.py) ... \u001b[?25l\u001b[?25hdone\n\u001b[31mERROR: tensorflow 2.5.0 has requirement tensorboard~=2.5, but you'll have tensorboard 2.4.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement pandas~=1.1.0; python_version >= \"3.0\", but you'll have pandas 1.3.0 which is incompatible.\u001b[0m\n\u001b[31mERROR: albumentations 0.1.12 has requirement imgaug<0.2.7,>=0.2.5, but you'll have imgaug 0.2.9 which is incompatible.\u001b[0m\n"
],
[
"from ttctext.datamodules.sst import SSTDataModule\nfrom ttctext.datasets.sst import StanfordSentimentTreeBank",
"_____no_output_____"
],
[
"sst_dataset = SSTDataModule(batch_size=128)\nsst_dataset.setup()",
"Cached Downloading: sst_dataset.zip\nDownloading...\nFrom: https://drive.google.com/uc?id=1urNi0Rtp9XkvkxxeKytjl1WoYNYUEoPI\nTo: /root/.cache/gdown/tmp0azbz08d/dl\n5.04MB [00:00, 197MB/s]\n"
],
[
"import pytorch_lightning as pl\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torchmetrics.functional import accuracy, precision, recall, confusion_matrix\nfrom sklearn.metrics import classification_report\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport pandas as pd\n\nsns.set()\n\nclass SSTModel(pl.LightningModule):\n\n def __init__(self, hparams, *args, **kwargs):\n super().__init__()\n\n self.save_hyperparameters(hparams)\n\n self.num_classes = self.hparams.output_dim\n\n self.embedding = nn.Embedding(self.hparams.input_dim, self.hparams.embedding_dim)\n\n self.lstm = nn.LSTM(\n self.hparams.embedding_dim, \n self.hparams.hidden_dim, \n num_layers=self.hparams.num_layers,\n dropout=self.hparams.dropout,\n batch_first=True\n )\n\n self.proj_layer = nn.Sequential(\n nn.Linear(self.hparams.hidden_dim, self.hparams.hidden_dim),\n nn.BatchNorm1d(self.hparams.hidden_dim),\n nn.ReLU(),\n nn.Dropout(self.hparams.dropout),\n )\n\n self.fc = nn.Linear(self.hparams.hidden_dim, self.num_classes)\n\n self.loss = nn.CrossEntropyLoss()\n\n def init_state(self, sequence_length):\n return (torch.zeros(self.hparams.num_layers, sequence_length, self.hparams.hidden_dim).to(self.device),\n torch.zeros(self.hparams.num_layers, sequence_length, self.hparams.hidden_dim).to(self.device))\n\n def forward(self, text, text_length, prev_state=None):\n\n # [batch size, sentence length] => [batch size, sentence len, embedding size]\n embedded = self.embedding(text)\n\n # packs the input for faster forward pass in RNN\n packed = torch.nn.utils.rnn.pack_padded_sequence(\n embedded, text_length.to('cpu'), \n enforce_sorted=False, \n batch_first=True\n )\n \n # [batch size sentence len, embedding size] => \n # output: [batch size, sentence len, hidden size]\n # hidden: [batch size, 1, hidden size]\n packed_output, curr_state = self.lstm(packed, prev_state)\n\n hidden_state, cell_state = curr_state\n\n # print('hidden state shape: ', hidden_state.shape)\n # print('cell')\n\n # unpack packed sequence\n # unpacked, unpacked_len = torch.nn.utils.rnn.pad_packed_sequence(packed_output, batch_first=True)\n\n # print('unpacked: ', unpacked.shape)\n\n # [batch size, sentence len, hidden size] => [batch size, num classes]\n # output = self.proj_layer(unpacked[:, -1])\n output = self.proj_layer(hidden_state[-1])\n\n # print('output shape: ', output.shape)\n\n output = self.fc(output)\n\n return output, curr_state\n\n def shared_step(self, batch, batch_idx):\n label, text, text_length = batch\n\n logits, in_state = self(text, text_length)\n \n loss = self.loss(logits, label)\n\n pred = torch.argmax(F.log_softmax(logits, dim=1), dim=1)\n acc = accuracy(pred, label)\n\n metric = {'loss': loss, 'acc': acc, 'pred': pred, 'label': label}\n\n return metric\n\n\n def training_step(self, batch, batch_idx):\n metrics = self.shared_step(batch, batch_idx)\n\n log_metrics = {'train_loss': metrics['loss'], 'train_acc': metrics['acc']}\n\n self.log_dict(log_metrics, prog_bar=True)\n\n return metrics\n\n\n def validation_step(self, batch, batch_idx):\n metrics = self.shared_step(batch, batch_idx)\n\n return metrics\n \n\n def validation_epoch_end(self, outputs):\n acc = torch.stack([x['acc'] for x in outputs]).mean()\n loss = torch.stack([x['loss'] for x in outputs]).mean()\n\n log_metrics = {'val_loss': loss, 'val_acc': acc}\n\n self.log_dict(log_metrics, prog_bar=True)\n\n if self.trainer.sanity_checking:\n return log_metrics\n\n preds = torch.cat([x['pred'] for x in outputs]).view(-1)\n labels = torch.cat([x['label'] for x in outputs]).view(-1)\n\n accuracy_ = accuracy(preds, labels)\n precision_ = precision(preds, labels, average='macro', num_classes=self.num_classes)\n recall_ = recall(preds, labels, average='macro', num_classes=self.num_classes) \n classification_report_ = classification_report(labels.cpu().numpy(), preds.cpu().numpy(), target_names=self.hparams.class_labels)\n confusion_matrix_ = confusion_matrix(preds, labels, num_classes=self.num_classes)\n cm_df = pd.DataFrame(confusion_matrix_.cpu().numpy(), index=self.hparams.class_labels, columns=self.hparams.class_labels)\n\n print(f'Test Epoch {self.current_epoch}/{self.hparams.epochs-1}: F1 Score: {accuracy_:.5f}, Precision: {precision_:.5f}, Recall: {recall_:.5f}\\n')\n print(f'Classification Report\\n{classification_report_}')\n\n fig, ax = plt.subplots(figsize=(10, 8))\n heatmap = sns.heatmap(cm_df, annot=True, ax=ax, fmt='d') # font size\n locs, labels = plt.xticks()\n plt.setp(labels, rotation=45)\n locs, labels = plt.yticks()\n plt.setp(labels, rotation=45)\n\n plt.show()\n\n print(\"\\n\")\n\n return log_metrics\n\n\n def test_step(self, batch, batch_idx):\n return self.validation_step(batch, batch_idx)\n\n def test_epoch_end(self, outputs):\n accuracy = torch.stack([x['acc'] for x in outputs]).mean()\n\n self.log('hp_metric', accuracy)\n\n self.log_dict({'test_acc': accuracy}, prog_bar=True)\n\n\n def configure_optimizers(self):\n optimizer = torch.optim.Adam(self.parameters(), lr=self.hparams.lr)\n lr_scheduler = {\n 'scheduler': torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, patience=10, verbose=True),\n 'monitor': 'train_loss',\n 'name': 'scheduler'\n }\n return [optimizer], [lr_scheduler]\n",
"_____no_output_____"
],
[
"from omegaconf import OmegaConf",
"_____no_output_____"
],
[
"hparams = OmegaConf.create({\n 'input_dim': len(sst_dataset.get_vocab()),\n 'embedding_dim': 128,\n 'num_layers': 2,\n 'hidden_dim': 64,\n 'dropout': 0.5,\n 'output_dim': len(StanfordSentimentTreeBank.get_labels()),\n 'class_labels': sst_dataset.raw_dataset_train.get_labels(),\n 'lr': 5e-4,\n 'epochs': 10,\n 'use_lr_finder': False\n})",
"_____no_output_____"
],
[
"sst_model = SSTModel(hparams)",
"_____no_output_____"
],
[
"trainer = pl.Trainer(gpus=1, max_epochs=hparams.epochs, progress_bar_refresh_rate=1, reload_dataloaders_every_epoch=True)",
"GPU available: True, used: True\nTPU available: False, using: 0 TPU cores\n"
],
[
"trainer.fit(sst_model, sst_dataset)",
"LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]\n\n | Name | Type | Params\n------------------------------------------------\n0 | embedding | Embedding | 3.1 M \n1 | lstm | LSTM | 82.9 K\n2 | proj_layer | Sequential | 4.3 K \n3 | fc | Linear | 325 \n4 | loss | CrossEntropyLoss | 0 \n------------------------------------------------\n3.2 M Trainable params\n0 Non-trainable params\n3.2 M Total params\n12.785 Total estimated model params size (MB)\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d00190b28512313d151ae3039c103cb50b48a002 | 377,328 | ipynb | Jupyter Notebook | make_a_better_business_decision_by_data/src/Analysis Feedback Data.ipynb | SteveZhengMe/devops-game-training | fe4ca362255562141df9cd9cd7e0e30c3efd8dec | [
"Apache-2.0"
] | null | null | null | make_a_better_business_decision_by_data/src/Analysis Feedback Data.ipynb | SteveZhengMe/devops-game-training | fe4ca362255562141df9cd9cd7e0e30c3efd8dec | [
"Apache-2.0"
] | null | null | null | make_a_better_business_decision_by_data/src/Analysis Feedback Data.ipynb | SteveZhengMe/devops-game-training | fe4ca362255562141df9cd9cd7e0e30c3efd8dec | [
"Apache-2.0"
] | null | null | null | 2,754.218978 | 364,917 | 0.621459 | [
[
[
"import lib.ana_header as lib\n\ndata = lib.AnalysisData()\n\ndata.sampleData()",
"_____no_output_____"
],
[
"result = data.categorize()\nresult",
"_____no_output_____"
],
[
"data.clusterPlot(result).show()",
"_____no_output_____"
],
[
"data.runSimulation(0)\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d0019907c02694279da2320dfc5ec1a80ea14989 | 84,706 | ipynb | Jupyter Notebook | examples/MultiGroupDirectLiNGAM.ipynb | YanaZeng/lingam | c16caf564c9f4e43eead65405189ab7ac2ae3f0d | [
"MIT"
] | 159 | 2019-08-22T05:17:19.000Z | 2022-03-28T23:41:27.000Z | examples/MultiGroupDirectLiNGAM.ipynb | YanaZeng/lingam | c16caf564c9f4e43eead65405189ab7ac2ae3f0d | [
"MIT"
] | 14 | 2020-04-26T17:25:42.000Z | 2022-02-14T08:05:05.000Z | examples/MultiGroupDirectLiNGAM.ipynb | YanaZeng/lingam | c16caf564c9f4e43eead65405189ab7ac2ae3f0d | [
"MIT"
] | 27 | 2020-01-19T07:31:08.000Z | 2021-12-26T06:23:35.000Z | 42.867409 | 7,748 | 0.510153 | [
[
[
"# MultiGroupDirectLiNGAM",
"_____no_output_____"
],
[
"## Import and settings\nIn this example, we need to import `numpy`, `pandas`, and `graphviz` in addition to `lingam`.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport graphviz\nimport lingam\nfrom lingam.utils import print_causal_directions, print_dagc, make_dot\n\nprint([np.__version__, pd.__version__, graphviz.__version__, lingam.__version__])\n\nnp.set_printoptions(precision=3, suppress=True)\nnp.random.seed(0)",
"['1.16.2', '0.24.2', '0.11.1', '1.5.4']\n"
]
],
[
[
"## Test data\nWe generate two datasets consisting of 6 variables.",
"_____no_output_____"
]
],
[
[
"x3 = np.random.uniform(size=1000)\nx0 = 3.0*x3 + np.random.uniform(size=1000)\nx2 = 6.0*x3 + np.random.uniform(size=1000)\nx1 = 3.0*x0 + 2.0*x2 + np.random.uniform(size=1000)\nx5 = 4.0*x0 + np.random.uniform(size=1000)\nx4 = 8.0*x0 - 1.0*x2 + np.random.uniform(size=1000)\nX1 = pd.DataFrame(np.array([x0, x1, x2, x3, x4, x5]).T ,columns=['x0', 'x1', 'x2', 'x3', 'x4', 'x5'])\nX1.head()",
"_____no_output_____"
],
[
"m = np.array([[0.0, 0.0, 0.0, 3.0, 0.0, 0.0],\n [3.0, 0.0, 2.0, 0.0, 0.0, 0.0],\n [0.0, 0.0, 0.0, 6.0, 0.0, 0.0],\n [0.0, 0.0, 0.0, 0.0, 0.0, 0.0],\n [8.0, 0.0,-1.0, 0.0, 0.0, 0.0],\n [4.0, 0.0, 0.0, 0.0, 0.0, 0.0]])\n\nmake_dot(m)",
"_____no_output_____"
],
[
"x3 = np.random.uniform(size=1000)\nx0 = 3.5*x3 + np.random.uniform(size=1000)\nx2 = 6.5*x3 + np.random.uniform(size=1000)\nx1 = 3.5*x0 + 2.5*x2 + np.random.uniform(size=1000)\nx5 = 4.5*x0 + np.random.uniform(size=1000)\nx4 = 8.5*x0 - 1.5*x2 + np.random.uniform(size=1000)\nX2 = pd.DataFrame(np.array([x0, x1, x2, x3, x4, x5]).T ,columns=['x0', 'x1', 'x2', 'x3', 'x4', 'x5'])\nX2.head()",
"_____no_output_____"
],
[
"m = np.array([[0.0, 0.0, 0.0, 3.5, 0.0, 0.0],\n [3.5, 0.0, 2.5, 0.0, 0.0, 0.0],\n [0.0, 0.0, 0.0, 6.5, 0.0, 0.0],\n [0.0, 0.0, 0.0, 0.0, 0.0, 0.0],\n [8.5, 0.0,-1.5, 0.0, 0.0, 0.0],\n [4.5, 0.0, 0.0, 0.0, 0.0, 0.0]])\n\nmake_dot(m)",
"_____no_output_____"
]
],
[
[
"We create a list variable that contains two datasets.",
"_____no_output_____"
]
],
[
[
"X_list = [X1, X2]",
"_____no_output_____"
]
],
[
[
"## Causal Discovery\nTo run causal discovery for multiple datasets, we create a `MultiGroupDirectLiNGAM` object and call the `fit` method.",
"_____no_output_____"
]
],
[
[
"model = lingam.MultiGroupDirectLiNGAM()\nmodel.fit(X_list)",
"_____no_output_____"
]
],
[
[
"Using the `causal_order_` properties, we can see the causal ordering as a result of the causal discovery.",
"_____no_output_____"
]
],
[
[
"model.causal_order_",
"_____no_output_____"
]
],
[
[
"Also, using the `adjacency_matrix_` properties, we can see the adjacency matrix as a result of the causal discovery. As you can see from the following, DAG in each dataset is correctly estimated.",
"_____no_output_____"
]
],
[
[
"print(model.adjacency_matrices_[0])\nmake_dot(model.adjacency_matrices_[0])",
"[[0. 0. 0. 3.006 0. 0. ]\n [2.873 0. 1.969 0. 0. 0. ]\n [0. 0. 0. 5.882 0. 0. ]\n [0. 0. 0. 0. 0. 0. ]\n [6.095 0. 0. 0. 0. 0. ]\n [3.967 0. 0. 0. 0. 0. ]]\n"
],
[
"print(model.adjacency_matrices_[1])\nmake_dot(model.adjacency_matrices_[1])",
"[[ 0. 0. 0. 3.483 0. 0. ]\n [ 3.516 0. 2.466 0.165 0. 0. ]\n [ 0. 0. 0. 6.383 0. 0. ]\n [ 0. 0. 0. 0. 0. 0. ]\n [ 8.456 0. -1.471 0. 0. 0. ]\n [ 4.446 0. 0. 0. 0. 0. ]]\n"
]
],
[
[
"To compare, we run DirectLiNGAM with single dataset concatenating two datasets.",
"_____no_output_____"
]
],
[
[
"X_all = pd.concat([X1, X2])\nprint(X_all.shape)",
"(2000, 6)\n"
],
[
"model_all = lingam.DirectLiNGAM()\nmodel_all.fit(X_all)\n\nmodel_all.causal_order_",
"_____no_output_____"
]
],
[
[
"You can see that the causal structure cannot be estimated correctly for a single dataset.",
"_____no_output_____"
]
],
[
[
"make_dot(model_all.adjacency_matrix_)",
"_____no_output_____"
]
],
[
[
"## Independence between error variables\nTo check if the LiNGAM assumption is broken, we can get p-values of independence between error variables. The value in the i-th row and j-th column of the obtained matrix shows the p-value of the independence of the error variables $e_i$ and $e_j$.",
"_____no_output_____"
]
],
[
[
"p_values = model.get_error_independence_p_values(X_list)\nprint(p_values[0])",
"[[0. 0.136 0.075 0.838 0. 0.832]\n [0.136 0. 0.008 0. 0.544 0.403]\n [0.075 0.008 0. 0.11 0. 0.511]\n [0.838 0. 0.11 0. 0.039 0.049]\n [0. 0.544 0. 0.039 0. 0.101]\n [0.832 0.403 0.511 0.049 0.101 0. ]]\n"
],
[
"print(p_values[1])",
"[[0. 0.545 0.908 0.285 0.525 0.728]\n [0.545 0. 0.84 0.814 0.086 0.297]\n [0.908 0.84 0. 0.032 0.328 0.026]\n [0.285 0.814 0.032 0. 0.904 0. ]\n [0.525 0.086 0.328 0.904 0. 0.237]\n [0.728 0.297 0.026 0. 0.237 0. ]]\n"
]
],
[
[
"## Bootstrapping\nIn `MultiGroupDirectLiNGAM`, bootstrap can be executed in the same way as normal `DirectLiNGAM`.",
"_____no_output_____"
]
],
[
[
"results = model.bootstrap(X_list, n_sampling=100)",
"_____no_output_____"
]
],
[
[
"## Causal Directions\nThe `bootstrap` method returns a list of multiple `BootstrapResult`, so we can get the result of bootstrapping from the list. We can get the same number of results as the number of datasets, so we specify an index when we access the results. We can get the ranking of the causal directions extracted by `get_causal_direction_counts()`.",
"_____no_output_____"
]
],
[
[
"cdc = results[0].get_causal_direction_counts(n_directions=8, min_causal_effect=0.01)\nprint_causal_directions(cdc, 100)",
"x0 <--- x3 (100.0%)\nx1 <--- x0 (100.0%)\nx1 <--- x2 (100.0%)\nx2 <--- x3 (100.0%)\nx4 <--- x0 (100.0%)\nx5 <--- x0 (100.0%)\nx4 <--- x2 (94.0%)\nx4 <--- x5 (20.0%)\n"
],
[
"cdc = results[1].get_causal_direction_counts(n_directions=8, min_causal_effect=0.01)\nprint_causal_directions(cdc, 100)",
"x0 <--- x3 (100.0%)\nx1 <--- x0 (100.0%)\nx1 <--- x2 (100.0%)\nx2 <--- x3 (100.0%)\nx4 <--- x0 (100.0%)\nx4 <--- x2 (100.0%)\nx5 <--- x0 (100.0%)\nx1 <--- x3 (72.0%)\n"
]
],
[
[
"## Directed Acyclic Graphs\nAlso, using the `get_directed_acyclic_graph_counts()` method, we can get the ranking of the DAGs extracted. In the following sample code, `n_dags` option is limited to the dags of the top 3 rankings, and `min_causal_effect` option is limited to causal directions with a coefficient of 0.01 or more.",
"_____no_output_____"
]
],
[
[
"dagc = results[0].get_directed_acyclic_graph_counts(n_dags=3, min_causal_effect=0.01)\nprint_dagc(dagc, 100)",
"DAG[0]: 61.0%\n\tx0 <--- x3 \n\tx1 <--- x0 \n\tx1 <--- x2 \n\tx2 <--- x3 \n\tx4 <--- x0 \n\tx4 <--- x2 \n\tx5 <--- x0 \nDAG[1]: 13.0%\n\tx0 <--- x3 \n\tx1 <--- x0 \n\tx1 <--- x2 \n\tx2 <--- x3 \n\tx4 <--- x0 \n\tx4 <--- x2 \n\tx4 <--- x5 \n\tx5 <--- x0 \nDAG[2]: 6.0%\n\tx0 <--- x3 \n\tx1 <--- x0 \n\tx1 <--- x2 \n\tx2 <--- x3 \n\tx4 <--- x0 \n\tx5 <--- x0 \n"
],
[
"dagc = results[1].get_directed_acyclic_graph_counts(n_dags=3, min_causal_effect=0.01)\nprint_dagc(dagc, 100)",
"DAG[0]: 59.0%\n\tx0 <--- x3 \n\tx1 <--- x0 \n\tx1 <--- x2 \n\tx1 <--- x3 \n\tx2 <--- x3 \n\tx4 <--- x0 \n\tx4 <--- x2 \n\tx5 <--- x0 \nDAG[1]: 17.0%\n\tx0 <--- x3 \n\tx1 <--- x0 \n\tx1 <--- x2 \n\tx2 <--- x3 \n\tx4 <--- x0 \n\tx4 <--- x2 \n\tx5 <--- x0 \nDAG[2]: 10.0%\n\tx0 <--- x2 \n\tx0 <--- x3 \n\tx1 <--- x0 \n\tx1 <--- x2 \n\tx1 <--- x3 \n\tx2 <--- x3 \n\tx4 <--- x0 \n\tx4 <--- x2 \n\tx5 <--- x0 \n"
]
],
[
[
"## Probability\nUsing the get_probabilities() method, we can get the probability of bootstrapping.",
"_____no_output_____"
]
],
[
[
"prob = results[0].get_probabilities(min_causal_effect=0.01)\nprint(prob)",
"[[0. 0. 0.08 1. 0. 0. ]\n [1. 0. 1. 0.08 0. 0.05]\n [0. 0. 0. 1. 0. 0. ]\n [0. 0. 0. 0. 0. 0. ]\n [1. 0. 0.94 0. 0. 0.2 ]\n [1. 0. 0. 0. 0.01 0. ]]\n"
]
],
[
[
"## Total Causal Effects\nUsing the `get_total_causal_effects()` method, we can get the list of total causal effect. The total causal effects we can get are dictionary type variable.\nWe can display the list nicely by assigning it to pandas.DataFrame. Also, we have replaced the variable index with a label below.",
"_____no_output_____"
]
],
[
[
"causal_effects = results[0].get_total_causal_effects(min_causal_effect=0.01)\ndf = pd.DataFrame(causal_effects)\n\nlabels = [f'x{i}' for i in range(X1.shape[1])]\ndf['from'] = df['from'].apply(lambda x : labels[x])\ndf['to'] = df['to'].apply(lambda x : labels[x])\ndf",
"_____no_output_____"
]
],
[
[
"We can easily perform sorting operations with pandas.DataFrame.",
"_____no_output_____"
]
],
[
[
"df.sort_values('effect', ascending=False).head()",
"_____no_output_____"
]
],
[
[
"And with pandas.DataFrame, we can easily filter by keywords. The following code extracts the causal direction towards x1.",
"_____no_output_____"
]
],
[
[
"df[df['to']=='x1'].head()",
"_____no_output_____"
]
],
[
[
"Because it holds the raw data of the total causal effect (the original data for calculating the median), it is possible to draw a histogram of the values of the causal effect, as shown below.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()\n%matplotlib inline\n\nfrom_index = 3\nto_index = 0\nplt.hist(results[0].total_effects_[:, to_index, from_index])",
"_____no_output_____"
]
],
[
[
"## Bootstrap Probability of Path\nUsing the `get_paths()` method, we can explore all paths from any variable to any variable and calculate the bootstrap probability for each path. The path will be output as an array of variable indices. For example, the array `[3, 0, 1]` shows the path from variable X3 through variable X0 to variable X1.",
"_____no_output_____"
]
],
[
[
"from_index = 3 # index of x3\nto_index = 1 # index of x0\n\npd.DataFrame(results[0].get_paths(from_index, to_index))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0019c78a544ab0773cdff60738644e98408fc67 | 414,096 | ipynb | Jupyter Notebook | 05_pandas.ipynb | stat4decision/python-data-ipsos-fev20 | a4b312989b8a2322b5aae80135446d33a9b8f7d1 | [
"MIT"
] | null | null | null | 05_pandas.ipynb | stat4decision/python-data-ipsos-fev20 | a4b312989b8a2322b5aae80135446d33a9b8f7d1 | [
"MIT"
] | null | null | null | 05_pandas.ipynb | stat4decision/python-data-ipsos-fev20 | a4b312989b8a2322b5aae80135446d33a9b8f7d1 | [
"MIT"
] | null | null | null | 77.285554 | 194,672 | 0.724557 | [
[
[
"\n\nPandas est le package de prédilection pour traiter des données structurées.\n\nPandas est basé sur 2 structures extrêmement liées les Series et le DataFrame.\n\nCes deux structures permettent de traiter des données sous forme de tableaux indexés.\n\nLes classes de Pandas utilisent des classes de Numpy, il est donc possible d'utiliser les fonctions universelles de Numpy sur les objets Pandas.",
"_____no_output_____"
]
],
[
[
"# on importe pandas avec :\nimport pandas as pd\nimport numpy as np\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# Les Series de Pandas\n\n- Les Series sont indexées, c'est leur avantage sur les arrays de NumPy\n- On peut utiliser les fonctions `.values` et `.index` pour voir les différentes parties de chaque Series\n- On définit une Series par `pd.Series([,], index=['','',])`\n- On peut appeler un élément avec `ma_serie['France']`\n- On peut aussi faire des conditions :\n```python\nma_serie[ma_serie>5000000]\n```\n```\n'France' in ma_serie\n```\n- Les objets Series peuvent être transformés en dictionnaires en utilisant :\n`.to_dict()`",
"_____no_output_____"
],
[
"**Exercice :**\n \nDéfinir un objet Series comprenant la population de 5 pays puis afficher les pays ayant une population > 50’000’000.\n",
"_____no_output_____"
]
],
[
[
"ser_pop = pd.Series([70,8,300,1200],index=[\"France\",\"Suisse\",\"USA\",\"Chine\"])",
"_____no_output_____"
],
[
"ser_pop",
"_____no_output_____"
],
[
"# on extrait une valeur avec une clé\nser_pop[\"France\"]",
"_____no_output_____"
],
[
"# on peut aussi utiliser une position avec .iloc[]\nser_pop.iloc[0]",
"_____no_output_____"
],
[
"# on applique la condition entre []\nser_pop[ser_pop>50]",
"_____no_output_____"
]
],
[
[
"# D'autres opérations sur les objets series\n\n- Pour définir le nom de la Series, on utilise `.name`\n- Pour définir le titre de la colonne des observations, on utilise `.index.name`",
"_____no_output_____"
],
[
"**Exercice :**\n \nDéfinir les noms de l’objet et de la colonne des pays pour la Series précédente\n",
"_____no_output_____"
]
],
[
[
"ser_pop.name = \"Populations\"\nser_pop.index.name = \"Pays\"",
"_____no_output_____"
],
[
"ser_pop",
"_____no_output_____"
]
],
[
[
"# Les données manquantes\n\nDans pandas, les données manquantes sont identifiés avec les fonctions de Numpy (`np.nan`). On a d'autres fonctions telles que :",
"_____no_output_____"
]
],
[
[
"pd.Series([2,np.nan,4],index=['a','b','c'])",
"_____no_output_____"
],
[
"pd.isna(pd.Series([2,np.nan,4],index=['a','b','c']))",
"_____no_output_____"
],
[
"pd.notna(pd.Series([2,np.nan,4],index=['a','b','c']))",
"_____no_output_____"
]
],
[
[
"# Les dates avec pandas\n\n- Python possède un module datetime qui permet de gérer facilement des dates\n- Pandas permet d'appliquer les opérations sur les dates aux Series et aux DataFrame\n- Le format es dates Python est `YYYY-MM-DD HH:MM:SS`\n\n- On peut générer des dates avec la fonction `pd.date_range()` avec différente fréquences `freq=`\n- On peut utiliser ces dates comme index dans un DataFrame ou dans un objet Series\n- On peut changer la fréquence en utilisant `.asfreq()`\n- Pour transformer une chaine de caractère en date, on utilise `pd.to_datetime()` avec l’option `dayfirst=True` si on est dans le cas français\n-On pourra aussi spécifier un format pour accélérer le processus `%Y%m%d`",
"_____no_output_____"
],
[
"**Exercice :**\n\nCréez un objet Series et ajoutez des dates partant du 3 octobre 2017 par jour jusqu’à aujourd’hui. Afficher le résultat dans un graphique (on utilisera la méthode `.plot()`",
"_____no_output_____"
]
],
[
[
"dates = pd.date_range(\"2017-10-03\", \"2020-02-27\",freq=\"W\")\n\nvaleurs = np.random.random(size=len(dates))\nma_serie=pd.Series(valeurs, index =dates)\nma_serie.plot()",
"_____no_output_____"
],
[
"len(dates)",
"_____no_output_____"
]
],
[
[
"# Le DataFrame \n\n- Les DataFrame sont des objets très souples pouvant être construits de différentes façon\n- On peut les construire en récupérant des données copier / coller, où directement sur Internet, ou en entrant les valeurs manuellement\n\n\n- Les DataFrame se rapprochent des dictionnaires et on peut construire ces objets en utilisant `DataFrame(dico)`\n- De nombreux détails sur la création des DataFrame se trouve sur ce site :\n\n<http://pandas.pydata.org/pandas-docs/dev/generated/pandas.DataFrame.html>\n",
"_____no_output_____"
],
[
"# Construction de DataFrame\n\nOn peut simplement construire un DataFrame avec le classe pd.DataFrame() à partir de différentes structures :",
"_____no_output_____"
]
],
[
[
"frame1=pd.DataFrame(np.random.randn(10).reshape(5,2),\n index=[\"obs_\"+str(i) for i in range(5)],\n columns=[\"col_\"+str(i) for i in range(2)])\nframe1",
"_____no_output_____"
]
],
[
[
"# Opérations sur les DataFrame\n\nOn peut afficher le nom des colonnes :",
"_____no_output_____"
]
],
[
[
"print(frame1.columns)",
"Index(['col_0', 'col_1'], dtype='object')\n"
]
],
[
[
"On peut accéder à une colonne avec :\n- `frame1.col_0` : attention au cas de nom de colonnes avec des espaces...\n- `frame1['col_0']`\n\nOn peut accéder à une cellule avec :\n- `frame1.loc['obs1','col_0']` : on utilise les index et le nom des colonnes\n- `frame1.iloc[1,0]` : on utilise les positions dans le DataFrame\n",
"_____no_output_____"
],
[
"# Options de visualisation et de résumé\n\nPour afficher les 3 premières lignes, on peut utiliser :\n",
"_____no_output_____"
]
],
[
[
"frame1.head(3)",
"_____no_output_____"
]
],
[
[
"Pour afficher un résumé du DF :",
"_____no_output_____"
]
],
[
[
"frame1.info()",
"<class 'pandas.core.frame.DataFrame'>\nIndex: 5 entries, obs_0 to obs_4\nData columns (total 2 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 col_0 5 non-null float64\n 1 col_1 5 non-null float64\ndtypes: float64(2)\nmemory usage: 120.0+ bytes\n"
]
],
[
[
"# Importer des données externes\n\nPandas est l'outil le plus efficace pour importer des données externes, il prend en charge de nombreux formats dont csv, Excel, SQL, SAS...\n\n\n## Importation de données avec Pandas\n\nQuel que soit le type de fichier, Pandas possède une fonction :\n```python\nframe=pd.read_...('chemin_du_fichier/nom_du_fichier',...)\n```\nPour écrire un DataFrame dans un fichier, on utilise :\n```python\nframe.to_...('chemin_du_fichier/nom_du_fichier',...)\n```",
"_____no_output_____"
],
[
"**Exercice :**\n \nImporter un fichier `.csv` avec `pd.read_csv()`. On utilisera le fichier \"./data/airbnb.csv\"",
"_____no_output_____"
]
],
[
[
"# on prend la colonne id comme index de notre DataFrame\nairbnb = pd.read_csv(\"https://www.stat4decision.com/airbnb.csv\",index_col=\"id\")",
"C:\\Users\\s4d-asus-14\\Anaconda3\\lib\\site-packages\\IPython\\core\\interactiveshell.py:3063: DtypeWarning: Columns (43,61,62) have mixed types.Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
],
[
"airbnb.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 59126 entries, 3109 to 34477345\nColumns: 105 entries, listing_url to reviews_per_month\ndtypes: float64(23), int64(20), object(62)\nmemory usage: 47.8+ MB\n"
],
[
"# la colonne price est sous forme d'objet et donc de chaîne de caractères\n# on a 2933 locations qui coûtent 80$ la nuit\nairbnb[\"price\"].value_counts()",
"_____no_output_____"
],
[
"dpt = pd.read_csv(\"./data/base-dpt.csv\", sep = \";\")\ndpt.head()",
"_____no_output_____"
],
[
"dpt.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1300 entries, 0 to 1299\nData columns (total 38 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 CODGEO 1300 non-null int64 \n 1 LIBGEO 1300 non-null object \n 2 REG 1300 non-null int64 \n 3 DEP 1300 non-null int64 \n 4 P14_POP 1297 non-null float64\n 5 P09_POP 1297 non-null float64\n 6 SUPERF 1297 non-null float64\n 7 NAIS0914 1297 non-null float64\n 8 DECE0914 1297 non-null float64\n 9 P14_MEN 1297 non-null float64\n 10 NAISD16 1296 non-null float64\n 11 DECESD16 1296 non-null float64\n 12 P14_LOG 1297 non-null float64\n 13 P14_RP 1297 non-null float64\n 14 P14_RSECOCC 1297 non-null float64\n 15 P14_LOGVAC 1297 non-null float64\n 16 P14_RP_PROP 1297 non-null float64\n 17 NBMENFISC14 1280 non-null float64\n 18 PIMP14 561 non-null float64\n 19 MED14 1280 non-null float64\n 20 TP6014 462 non-null float64\n 21 P14_EMPLT 1297 non-null float64\n 22 P14_EMPLT_SAL 1297 non-null float64\n 23 P09_EMPLT 1300 non-null float64\n 24 P14_POP1564 1297 non-null float64\n 25 P14_CHOM1564 1297 non-null float64\n 26 P14_ACT1564 1297 non-null float64\n 27 ETTOT15 1299 non-null float64\n 28 ETAZ15 1299 non-null float64\n 29 ETBE15 1299 non-null float64\n 30 ETFZ15 1299 non-null float64\n 31 ETGU15 1299 non-null float64\n 32 ETGZ15 1299 non-null float64\n 33 ETOQ15 1299 non-null float64\n 34 ETTEF115 1299 non-null float64\n 35 ETTEFP1015 1299 non-null float64\n 36 Geo Shape 1297 non-null object \n 37 geo_point_2d 1297 non-null object \ndtypes: float64(32), int64(3), object(3)\nmemory usage: 386.1+ KB\n"
]
],
[
[
"# D'autres types de données\n\n## JSON\nLes objets JSON ressemblent à des dictionnaires.\n\nOn utilise le module `json` puis la fonction `json.loads()` pour transformer une entrée JSON en objet json\n\n## HTML\nOn utilise `pd.read_html(url)`. Cet fonction est basée sur les packages `beautifulsoup` et `html5lib`\n\nCette fonction renvoie une liste de DataFrame qui représentent tous les DataFrame de la page. On ira ensuite chercher l'élément qui nous intéresse avec `frame_list[0]`",
"_____no_output_____"
],
[
"**Exercice :**\n \nImportez un tableau en html depuis la page <http://www.fdic.gov/bank/individual/failed/banklist.html>",
"_____no_output_____"
]
],
[
[
"bank = pd.read_html(\"http://www.fdic.gov/bank/individual/failed/banklist.html\")\n# read_html() stocke les tableaux d'une page web dans une liste\ntype(bank)",
"_____no_output_____"
],
[
"len(bank)",
"_____no_output_____"
],
[
"bank[0].head(10)",
"_____no_output_____"
],
[
"nba = pd.read_html(\"https://en.wikipedia.org/wiki/2018%E2%80%9319_NBA_season\")",
"_____no_output_____"
],
[
"len(nba)",
"_____no_output_____"
],
[
"nba[3]",
"_____no_output_____"
]
],
[
[
"# Importer depuis Excel\n\nOn a deux approches pour Excel :\n- On peut utiliser `pd.read_excel()`\n- On peut utiliser la classe `pd.ExcelFile()`\n\nDans ce cas, on utilise :\n```python\nxlsfile=pd.ExcelFile('fichier.xlsx')\nxlsfile.parse('Sheet1')\n```",
"_____no_output_____"
],
[
"**Exercice :** \n \nImportez un fichier Excel avec les deux approches, on utilisera : `credit2.xlsx` et `ville.xls`",
"_____no_output_____"
]
],
[
[
"pd.read_excel(\"./data/credit2.xlsx\",usecols=[\"Age\",\"Gender\"])\npd.read_excel(\"./data/credit2.xlsx\",usecols=\"A:C\")\ncredit2 = pd.read_excel(\"./data/credit2.xlsx\", index_col=\"Customer_ID\")",
"_____no_output_____"
],
[
"credit2.head()",
"_____no_output_____"
],
[
"# on crée un objet du type ExcelFile\nville = pd.ExcelFile(\"./data/ville.xls\")",
"_____no_output_____"
],
[
"ville.sheet_names",
"_____no_output_____"
],
[
"# on extrait toutes les feuilles avec le mot ville dans le nom de la feuille dans une liste de dataframes\nlist_feuilles_ville = []\nfor nom in ville.sheet_names:\n if \"ville\" in nom:\n list_feuilles_ville.append(ville.parse(nom))",
"_____no_output_____"
]
],
[
[
"On crée une fonction qui permet d'importer les feuilles excel ayant le terme nom_dans_feuille dans le nom de la feuille",
"_____no_output_____"
]
],
[
[
"def import_excel_feuille(chemin_fichier, nom_dans_feuille = \"\"):\n \"\"\" fonction qui importe les feuilles excel ayant le terme nom_dans_feuille dans le nom de la feuille\"\"\"\n \n excel = pd.ExcelFile(chemin_fichier)\n list_feuilles = []\n for nom_feuille in excel.sheet_names:\n if nom_dans_feuille in nom_feuille:\n list_feuilles.append(excel.parse(nom))\n return list_feuilles",
"_____no_output_____"
],
[
"list_ain = import_excel_feuille(\"./data/ville.xls\",nom_dans_feuille=\"ain\")",
"_____no_output_____"
],
[
"list_ain[0].head()",
"_____no_output_____"
]
],
[
[
"# Importer des données SQL\n\nPandas possède une fonction `read_sql()` qui permet d’importer directement des bases de données ou des queries dans des DataFrame\n\nIl faut tout de même un connecteur pour accéder aux bases de données\n\nPour mettre en place ce connecteur, on utlise le package SQLAlchemy.\n\nSuivant le type de base de données, on utilisera différents codes mais la structure du code est toujours la même",
"_____no_output_____"
]
],
[
[
"# on importe l'outil de connexion\nfrom sqlalchemy import create_engine",
"_____no_output_____"
]
],
[
[
"On crée une connexion\n```python\nconnexion=create_engine(\"sqlite:///(...).sqlite\")\n```",
"_____no_output_____"
],
[
"On utlise une des fonctions de Pandas pour charger les données\n```python\nrequete=\"\"\"select ... from ...\"\"\"\nframe_sql=pd.read_sql_query(requete,connexion)\n```",
"_____no_output_____"
],
[
"**Exercices :**\n \nImportez la base de données SQLite salaries et récupérez la table Salaries dans un DataFrame ",
"_____no_output_____"
]
],
[
[
"connexion=create_engine(\"sqlite:///./data/salaries.sqlite\")",
"_____no_output_____"
],
[
"connexion.table_names()",
"_____no_output_____"
],
[
"salaries = pd.read_sql_query(\"select * from salaries\", con=connexion)",
"_____no_output_____"
],
[
"salaries.head()",
"_____no_output_____"
]
],
[
[
"# Importer depuis SPSS\n\nPandas possède une fonction `pd.read_spss()`\n\nAttention ! Il faut la dernière version de Pandas et installer des packages supplémentaires !\n\n**Exercice :** Importer le fichier SPSS se trouvant dans ./data/",
"_____no_output_____"
]
],
[
[
"#base = pd.read_spss(\"./data/Base.sav\")",
"_____no_output_____"
]
],
[
[
"# Les tris avec Pandas \n\nPour effectuer des tris, on utilise :\n- `.sort_index()` pour le tri des index\n- `.sort_values()` pour le tri des données\n- `.rank()` affiche le rang des observations\n\nIl peut y avoir plusieurs tris dans la même opération. Dans ce cas, on utilise des listes de colonnes :\n```python\nframe.sort_values([\"col_1\",\"col_2\"])\n```",
"_____no_output_____"
],
[
"**Exercice :** \n \nTriez les données sur les salaires en se basant sur le TotalPay et le JobTitle",
"_____no_output_____"
]
],
[
[
"salaries.sort_values([\"JobTitle\",\"TotalPay\"],ascending=[True, False])",
"_____no_output_____"
]
],
[
[
"# Les statistiques simples\n\nLes Dataframe possèdent de nombreuses méthodes pour calculer des statistiques simples :\n- `.sum(axis=0)` permet de faire une somme par colonne\n- `.sum(axis=1)` permet de faire une somme par ligne\n- `.min()` et `.max()` donnent le minimum par colonne\n- `.idxmin()` et `.idxmax()` donnent l’index du minimum et du maximum\n- `.describe()` affiche un tableau de statistiques descriptives par colonne\n- `.corr()` pour calculer la corrélation entre les colonnes",
"_____no_output_____"
],
[
"**Exercice :**\n \nObtenir les différentes statistiques descriptives pour les données AirBnB.\n\nOn peut s'intéresser à la colonne `Price` (attention des prétraitements sont nécessaires)\n",
"_____no_output_____"
]
],
[
[
"# cette colonne est sous forme d'object, il va falloir la modifier\nairbnb[\"price\"].dtype",
"_____no_output_____"
],
[
"airbnb[\"price_num\"] = pd.to_numeric(airbnb[\"price\"].str.replace(\"$\",\"\")\n .str.replace(\",\",\"\"))",
"_____no_output_____"
],
[
"airbnb[\"price_num\"].dtype",
"_____no_output_____"
],
[
"airbnb[\"price_num\"].mean()",
"_____no_output_____"
],
[
"airbnb[\"price_num\"].describe()",
"_____no_output_____"
],
[
"# on extrait l'id de la location avec le prix max\nairbnb[\"price_num\"].idxmax()",
"_____no_output_____"
],
[
"# on affiche cette location\nairbnb.loc[airbnb[\"price_num\"].idxmax()]",
"_____no_output_____"
]
],
[
[
"Calcul de la moyenne pondérée sur une enquête",
"_____no_output_____"
]
],
[
[
"base = pd.read_csv(\"./data/Base.csv\")",
"_____no_output_____"
],
[
"#moyenne pondérée\nnp.average(base[\"resp_age\"],weights=base[\"Weight\"])",
"_____no_output_____"
],
[
"# moyenne\nbase[\"resp_age\"].mean()",
"_____no_output_____"
]
],
[
[
"Utilisation de statsmodels",
"_____no_output_____"
]
],
[
[
"from statsmodels.stats.weightstats import DescrStatsW",
"C:\\Users\\s4d-asus-14\\Anaconda3\\lib\\site-packages\\statsmodels\\tools\\_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
],
[
"# on sélectionne les colonnes numériques\nbase_num = base.select_dtypes(np.number)\n# on calcule les stats desc pondérées\nmes_stat = DescrStatsW(base_num, weights=base[\"Weight\"])",
"_____no_output_____"
],
[
"base_num.columns",
"_____no_output_____"
],
[
"mes_stat.var",
"_____no_output_____"
],
[
"mes_stat_age = DescrStatsW(base[\"resp_age\"], weights=base[\"Weight\"])",
"_____no_output_____"
],
[
"mes_stat_age.mean",
"_____no_output_____"
]
],
[
[
"On va construire une fonction permettant de calculer les stat desc pondérées d'une colonne",
"_____no_output_____"
]
],
[
[
"def stat_desc_w_ipsos(data, columns, weights):\n \"\"\" Cette fonction calcule et affiche les moyennes et écarts-types pondérés\n \n Input : - data : données sous forme de DataFrame\n - columns : nom des colonnes quanti à analyser\n - weights : nom de la colonne des poids\n \"\"\"\n \n from statsmodels.stats.weightstats import DescrStatsW\n mes_stats = DescrStatsW(data[columns],weights=data[weights])\n print(\"Moyenne pondérée :\", mes_stats.mean)\n print(\"Ecart-type pondéré :\", mes_stats.std)\n ",
"_____no_output_____"
],
[
"stat_desc_w_ipsos(base,\"resp_age\",\"Weight\")",
"Moyenne pondérée : 48.40297631233564\nEcart-type pondéré : 17.1309963999935\n"
]
],
[
[
"# Le traitement des données manquantes\n\n- Les données manquantes sont identifiées par `NaN`\n\n\n- `.dropna()` permet de retirer les données manquantes dans un objet Series et l’ensemble d’une ligne dans le cas d’un DataFrame\n- Pour éliminer par colonne, on utilise `.dropna(axis=1)`\n- Remplacer toutes les données manquantes `.fillna(valeur)`\n",
"_____no_output_____"
],
[
"# Les jointures avec Pandas\n\nOn veut joindre des jeux de données en utilisant des clés (variables communes)\n\n- `pd.merge()` permet de joindre deux DataFrame, on utilise comme options `on='key'`\n\n- On peut utiliser comme option `how=`, on peut avoir :\n - `left` dans ce cas, on garde le jeu de données à gauche et pour les données de droite des valeurs manquantes sont ajoutées.\n - `outer`, on garde toutes les valeurs des deux jeux de données\n - ...\n\n- On peut avoir plusieurs clés et faire une jointure sur les deux clés `on=['key1','key2']`\n\nPour plus de détails : <http://pandas.pydata.org/pandas-docs/dev/generated/pandas.DataFrame.merge.html>\n",
"_____no_output_____"
],
[
"**Exercice :**\n \nJoindre deux dataframes (credit1 et credit2).\n",
"_____no_output_____"
]
],
[
[
"credit1 = pd.read_csv(\"./data/credit1.txt\",sep=\"\\t\")",
"_____no_output_____"
],
[
"credit_global = pd.merge(credit1,credit2,how=\"inner\",on=\"Customer_ID\")",
"_____no_output_____"
],
[
"credit_global.head()",
"_____no_output_____"
]
],
[
[
"On fait une jointure entre les données des locations Airbnb et les données de calendrier de remplissage des appartements",
"_____no_output_____"
]
],
[
[
"airbnb_reduit = airbnb[[\"price_num\",\"latitude\",\"longitude\"]]",
"_____no_output_____"
],
[
"calendar = pd.read_csv(\"https://www.stat4decision.com/calendar.csv.gz\")",
"_____no_output_____"
],
[
"calendar.head()",
"_____no_output_____"
],
[
"new_airbnb = pd.merge(calendar,airbnb[[\"price_num\",\"latitude\",\"longitude\"]], \n left_on = \"listing_id\",right_index=True)",
"_____no_output_____"
],
[
"new_airbnb.shape",
"_____no_output_____"
]
],
[
[
"On veut extraire des statistiques de base\n\nPar exemple, la moyenne des prix pour les locations du 8 juillet 2018 :",
"_____no_output_____"
]
],
[
[
"new_airbnb[new_airbnb[\"date\"]=='2018-07-08'][\"price_num\"].mean()",
"_____no_output_____"
]
],
[
[
"On extrait le nombre de nuitées disponibles / occuppées :",
"_____no_output_____"
]
],
[
[
"new_airbnb[\"available\"].value_counts(normalize = True)",
"_____no_output_____"
]
],
[
[
"Si on regarde le part de locations occuppées le 8 janvier 2019, on a :",
"_____no_output_____"
]
],
[
[
"new_airbnb[new_airbnb[\"date\"]=='2019-01-08'][\"available\"].value_counts(normalize = True)",
"_____no_output_____"
]
],
[
[
"La moyenne des prix des appartements disponibles le 8 juillet 2018 :",
"_____no_output_____"
]
],
[
[
"new_airbnb[(new_airbnb[\"date\"]=='2018-07-08')&(new_airbnb[\"available\"]=='t')][\"price_num\"].mean()",
"_____no_output_____"
]
],
[
[
"On transforme la colonne date qui est sous forme de chaîne de caractère en DateTime, ceci permet de faire de nouvelles opérations : ",
"_____no_output_____"
]
],
[
[
"new_airbnb[\"date\"]= pd.to_datetime(new_airbnb[\"date\"])",
"_____no_output_____"
],
[
"# on construit une colonne avec le jour de la semaine \nnew_airbnb[\"jour_semaine\"]=new_airbnb[\"date\"].dt.day_name()",
"_____no_output_____"
]
],
[
[
"La moyenne des pris des Samedi soirs disponibles est donc :",
"_____no_output_____"
]
],
[
[
"new_airbnb[(new_airbnb[\"jour_semaine\"]=='Saturday')&(new_airbnb[\"available\"]=='t')][\"price_num\"].mean()",
"_____no_output_____"
]
],
[
[
"# Gestion des duplications\n\n- On utilise `.duplicated()` ou `.drop_duplicates()` dans le cas où on désire effacer les lignes se répétant\n\n\n- On peut se concentrer sur une seule variables en entrant directement le nom de la variable. Dans ce cas, c’est la première apparition qui compte. Si on veut prendre la dernière apparition, on utilise l’option `keep=\"last\"`. On pourra avoir :\n```python\nframe1.drop_duplicates([\"col_0\",\"col_1\"],keep=\"last\")\n```",
"_____no_output_____"
],
[
"# Discrétisation\n\nPour discrétiser, on utilise la fonction `pd.cut()`, on va définir une liste de points pour discrétiser et on entre cette liste comme second paramètre de la fonction.\n\nUne fois discrétisé, on peut afficher les modalités obtenues en utilisant `.categories`\n\nOn peut aussi compter les occurrence en utilisant `pd.value_counts()`\n\nIl est aussi possible d’entrer le nombre de segments comme second paramètre\n\nOn utilisera aussi `qcut()`",
"_____no_output_____"
],
[
"**Exercice :**\n \nCréez une variable dans le dataframe AirBnB pour obtenir des niveaux de prix.\n",
"_____no_output_____"
]
],
[
[
"airbnb[\"price_disc1\"]=pd.cut(airbnb[\"price_num\"],bins=5)\nairbnb[\"price_disc2\"]=pd.qcut(airbnb[\"price_num\"],5)",
"_____no_output_____"
],
[
"airbnb[\"price_disc1\"].value_counts()",
"_____no_output_____"
],
[
"airbnb[\"price_disc2\"].value_counts()",
"_____no_output_____"
]
],
[
[
"# Les tableaux croisés avec Pandas\n\nLes DataFrame possèdent des méthodes pour générer des tableaux croisés, notamment :\n```python\nframe1.pivot_table()\n```\nCette méthode permet de gérer de nombreux cas avec des fonctions standards et sur mesure.",
"_____no_output_____"
],
[
"**Exercice :**\n \nAfficher un tableau Pivot pour les données AirBnB.",
"_____no_output_____"
]
],
[
[
"# on définit un \ndef moy2(x):\n return x.mean()/x.var()",
"_____no_output_____"
]
],
[
[
"On croise le room_type avec le niveau de prix et on regarde le review_score_rating moyen + le nombre d'occurences et une fonction \"maison\" :",
"_____no_output_____"
]
],
[
[
"airbnb['room_type']\nairbnb['price_disc2']\nairbnb['review_scores_rating']\nairbnb.pivot_table(values=[\"review_scores_rating\",'review_scores_cleanliness'],\n index=\"room_type\",\n columns='price_disc2',aggfunc=[\"count\",\"mean\",moy2])",
"_____no_output_____"
]
],
[
[
"# L'utilisation de GroupBy sur des DataFrame\n\n- `.groupby` permet de rassembler des observations en fonction d’une variable dite de groupe\n\n\n- Par exemple, `frame.groupby('X').mean()` donnera les moyennes par groupes de `X`\n\n\n- On peut aussi utiliser `.size()` pour connaître la taille des groupes et utiliser d’autres fonctions (`.sum()`)\n\n\n- On peut effectuer de nombreuses opérations de traitement avec le groupby\n",
"_____no_output_____"
]
],
[
[
"airbnb_group_room = airbnb.groupby(['room_type','price_disc2'])\n\nairbnb_group_room[\"price_num\"].describe()",
"_____no_output_____"
],
[
"# on peut afficher plusieurs statistiques\nairbnb_group_room[\"price_num\"].agg([\"mean\",\"median\",\"std\",\"count\"])",
"_____no_output_____"
],
[
"new_airbnb.groupby(['available','jour_semaine'])[\"price_num\"].agg([\"mean\",\"count\"])",
"_____no_output_____"
]
],
[
[
"Essayez d'utiliser une fonction lambda sur le groupby",
"_____no_output_____"
],
[
"**Exercice :**\n \n- Données sur les salaires\n\n\n- On utilise le `groupby()` pour rassembler les types d’emploi\n\n\n- Et on calcule des statistiques pour chaque type\n\n\nOn peut utiliser la méthode `.agg()` avec par exemple `'mean'` comme paramètre\n\nOn utilise aussi fréquemment la méthode `.apply()` combinée à une fonction lambda",
"_____no_output_____"
]
],
[
[
"# on passe tous les JobTitle en minuscule\nsalaries[\"JobTitle\"]= salaries[\"JobTitle\"].str.lower()",
"_____no_output_____"
],
[
"# nombre de JobTitle différents\nsalaries[\"JobTitle\"].nunique()",
"_____no_output_____"
],
[
"salaries.groupby(\"JobTitle\")[\"TotalPay\"].mean().sort_values(ascending=False)",
"_____no_output_____"
],
[
"salaries.groupby(\"JobTitle\")[\"TotalPay\"].agg([\"mean\",\"count\"]).sort_values(\"count\",ascending=False)",
"_____no_output_____"
]
],
[
[
"On peut aussi faire des représentations graphiques avancées :",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,5))\nplt.scatter(\"longitude\",\"latitude\", data = airbnb[airbnb[\"price_num\"]<150], s=1,c = \"price_num\", cmap=plt.get_cmap(\"jet\"))\nplt.colorbar()\nplt.savefig(\"paris_airbnb.jpg\")",
"_____no_output_____"
],
[
"airbnb[airbnb[\"price_num\"]<150]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d001a4f90a6a0c737eb8fb6f60a894a652aa8fe2 | 34,206 | ipynb | Jupyter Notebook | ml/cc/exercises/linear_regression_with_a_real_dataset.ipynb | lc0/eng-edu | 826da7f04e9dbde6bef08db3344ad385f3addcd6 | [
"Apache-2.0"
] | null | null | null | ml/cc/exercises/linear_regression_with_a_real_dataset.ipynb | lc0/eng-edu | 826da7f04e9dbde6bef08db3344ad385f3addcd6 | [
"Apache-2.0"
] | null | null | null | ml/cc/exercises/linear_regression_with_a_real_dataset.ipynb | lc0/eng-edu | 826da7f04e9dbde6bef08db3344ad385f3addcd6 | [
"Apache-2.0"
] | null | null | null | 38.870455 | 512 | 0.55005 | [
[
[
"#@title Copyright 2020 Google LLC. Double-click here for license information.\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Linear Regression with a Real Dataset\n\nThis Colab uses a real dataset to predict the prices of houses in California. \n\n\n\n\n",
"_____no_output_____"
],
[
"## Learning Objectives:\n\nAfter doing this Colab, you'll know how to do the following:\n\n * Read a .csv file into a [pandas](https://developers.google.com/machine-learning/glossary/#pandas) DataFrame.\n * Examine a [dataset](https://developers.google.com/machine-learning/glossary/#data_set). \n * Experiment with different [features](https://developers.google.com/machine-learning/glossary/#feature) in building a model.\n * Tune the model's [hyperparameters](https://developers.google.com/machine-learning/glossary/#hyperparameter).",
"_____no_output_____"
],
[
"## The Dataset\n \nThe [dataset for this exercise](https://developers.google.com/machine-learning/crash-course/california-housing-data-description) is based on 1990 census data from California. The dataset is old but still provides a great opportunity to learn about machine learning programming.",
"_____no_output_____"
],
[
"## Use the right version of TensorFlow\n\nThe following hidden code cell ensures that the Colab will run on TensorFlow 2.X.",
"_____no_output_____"
]
],
[
[
"#@title Run on TensorFlow 2.x\n%tensorflow_version 2.x",
"_____no_output_____"
]
],
[
[
"## Import relevant modules\n\nThe following hidden code cell imports the necessary code to run the code in the rest of this Colaboratory.",
"_____no_output_____"
]
],
[
[
"#@title Import relevant modules\nimport pandas as pd\nimport tensorflow as tf\nfrom matplotlib import pyplot as plt\n\n# The following lines adjust the granularity of reporting. \npd.options.display.max_rows = 10\npd.options.display.float_format = \"{:.1f}\".format",
"_____no_output_____"
]
],
[
[
"## The dataset\n\nDatasets are often stored on disk or at a URL in [.csv format](https://wikipedia.org/wiki/Comma-separated_values). \n\nA well-formed .csv file contains column names in the first row, followed by many rows of data. A comma divides each value in each row. For example, here are the first five rows of the .csv file file holding the California Housing Dataset:\n\n```\n\"longitude\",\"latitude\",\"housing_median_age\",\"total_rooms\",\"total_bedrooms\",\"population\",\"households\",\"median_income\",\"median_house_value\"\n-114.310000,34.190000,15.000000,5612.000000,1283.000000,1015.000000,472.000000,1.493600,66900.000000\n-114.470000,34.400000,19.000000,7650.000000,1901.000000,1129.000000,463.000000,1.820000,80100.000000\n-114.560000,33.690000,17.000000,720.000000,174.000000,333.000000,117.000000,1.650900,85700.000000\n-114.570000,33.640000,14.000000,1501.000000,337.000000,515.000000,226.000000,3.191700,73400.000000\n```\n\n",
"_____no_output_____"
],
[
"### Load the .csv file into a pandas DataFrame\n\nThis Colab, like many machine learning programs, gathers the .csv file and stores the data in memory as a pandas Dataframe. pandas is an open source Python library. The primary datatype in pandas is a DataFrame. You can imagine a pandas DataFrame as a spreadsheet in which each row is identified by a number and each column by a name. pandas is itself built on another open source Python library called NumPy. If you aren't familiar with these technologies, please view these two quick tutorials:\n\n* [NumPy](https://colab.research.google.com/github/google/eng-edu/blob/master/ml/cc/exercises/numpy_ultraquick_tutorial.ipynb?utm_source=linearregressionreal-colab&utm_medium=colab&utm_campaign=colab-external&utm_content=numpy_tf2-colab&hl=en)\n* [Pandas DataFrames](https://colab.research.google.com/github/google/eng-edu/blob/master/ml/cc/exercises/pandas_dataframe_ultraquick_tutorial.ipynb?utm_source=linearregressionreal-colab&utm_medium=colab&utm_campaign=colab-external&utm_content=pandas_tf2-colab&hl=en)\n\nThe following code cell imports the .csv file into a pandas DataFrame and scales the values in the label (`median_house_value`):",
"_____no_output_____"
]
],
[
[
"# Import the dataset.\ntraining_df = pd.read_csv(filepath_or_buffer=\"https://download.mlcc.google.com/mledu-datasets/california_housing_train.csv\")\n\n# Scale the label.\ntraining_df[\"median_house_value\"] /= 1000.0\n\n# Print the first rows of the pandas DataFrame.\ntraining_df.head()",
"_____no_output_____"
]
],
[
[
"Scaling `median_house_value` puts the value of each house in units of thousands. Scaling will keep loss values and learning rates in a friendlier range. \n\nAlthough scaling a label is usually *not* essential, scaling features in a multi-feature model usually *is* essential.",
"_____no_output_____"
],
[
"## Examine the dataset\n\nA large part of most machine learning projects is getting to know your data. The pandas API provides a `describe` function that outputs the following statistics about every column in the DataFrame:\n\n* `count`, which is the number of rows in that column. Ideally, `count` contains the same value for every column. \n\n* `mean` and `std`, which contain the mean and standard deviation of the values in each column. \n\n* `min` and `max`, which contain the lowest and highest values in each column.\n\n* `25%`, `50%`, `75%`, which contain various [quantiles](https://developers.google.com/machine-learning/glossary/#quantile).",
"_____no_output_____"
]
],
[
[
"# Get statistics on the dataset.\ntraining_df.describe()\n",
"_____no_output_____"
]
],
[
[
"### Task 1: Identify anomalies in the dataset\n\nDo you see any anomalies (strange values) in the data? ",
"_____no_output_____"
]
],
[
[
"#@title Double-click to view a possible answer.\n\n# The maximum value (max) of several columns seems very\n# high compared to the other quantiles. For example,\n# example the total_rooms column. Given the quantile\n# values (25%, 50%, and 75%), you might expect the \n# max value of total_rooms to be approximately \n# 5,000 or possibly 10,000. However, the max value \n# is actually 37,937.\n\n# When you see anomalies in a column, become more careful\n# about using that column as a feature. That said,\n# anomalies in potential features sometimes mirror \n# anomalies in the label, which could make the column \n# be (or seem to be) a powerful feature.\n# Also, as you will see later in the course, you \n# might be able to represent (pre-process) raw data \n# in order to make columns into useful features.",
"_____no_output_____"
]
],
[
[
"## Define functions that build and train a model\n\nThe following code defines two functions:\n\n * `build_model(my_learning_rate)`, which builds a randomly-initialized model.\n * `train_model(model, feature, label, epochs)`, which trains the model from the examples (feature and label) you pass. \n\nSince you don't need to understand model building code right now, we've hidden this code cell. You may optionally double-click the following headline to see the code that builds and trains a model.",
"_____no_output_____"
]
],
[
[
"#@title Define the functions that build and train a model\ndef build_model(my_learning_rate):\n \"\"\"Create and compile a simple linear regression model.\"\"\"\n # Most simple tf.keras models are sequential.\n model = tf.keras.models.Sequential()\n\n # Describe the topography of the model.\n # The topography of a simple linear regression model\n # is a single node in a single layer.\n model.add(tf.keras.layers.Dense(units=1, \n input_shape=(1,)))\n\n # Compile the model topography into code that TensorFlow can efficiently\n # execute. Configure training to minimize the model's mean squared error. \n model.compile(optimizer=tf.keras.optimizers.RMSprop(lr=my_learning_rate),\n loss=\"mean_squared_error\",\n metrics=[tf.keras.metrics.RootMeanSquaredError()])\n\n return model \n\n\ndef train_model(model, df, feature, label, epochs, batch_size):\n \"\"\"Train the model by feeding it data.\"\"\"\n\n # Feed the model the feature and the label.\n # The model will train for the specified number of epochs. \n history = model.fit(x=df[feature],\n y=df[label],\n batch_size=batch_size,\n epochs=epochs)\n\n # Gather the trained model's weight and bias.\n trained_weight = model.get_weights()[0]\n trained_bias = model.get_weights()[1]\n\n # The list of epochs is stored separately from the rest of history.\n epochs = history.epoch\n \n # Isolate the error for each epoch.\n hist = pd.DataFrame(history.history)\n\n # To track the progression of training, we're going to take a snapshot\n # of the model's root mean squared error at each epoch. \n rmse = hist[\"root_mean_squared_error\"]\n\n return trained_weight, trained_bias, epochs, rmse\n\nprint(\"Defined the create_model and traing_model functions.\")",
"_____no_output_____"
]
],
[
[
"## Define plotting functions\n\nThe following [matplotlib](https://developers.google.com/machine-learning/glossary/#matplotlib) functions create the following plots:\n\n* a scatter plot of the feature vs. the label, and a line showing the output of the trained model\n* a loss curve\n\nYou may optionally double-click the headline to see the matplotlib code, but note that writing matplotlib code is not an important part of learning ML programming.",
"_____no_output_____"
]
],
[
[
"#@title Define the plotting functions\ndef plot_the_model(trained_weight, trained_bias, feature, label):\n \"\"\"Plot the trained model against 200 random training examples.\"\"\"\n\n # Label the axes.\n plt.xlabel(feature)\n plt.ylabel(label)\n\n # Create a scatter plot from 200 random points of the dataset.\n random_examples = training_df.sample(n=200)\n plt.scatter(random_examples[feature], random_examples[label])\n\n # Create a red line representing the model. The red line starts\n # at coordinates (x0, y0) and ends at coordinates (x1, y1).\n x0 = 0\n y0 = trained_bias\n x1 = 10000\n y1 = trained_bias + (trained_weight * x1)\n plt.plot([x0, x1], [y0, y1], c='r')\n\n # Render the scatter plot and the red line.\n plt.show()\n\n\ndef plot_the_loss_curve(epochs, rmse):\n \"\"\"Plot a curve of loss vs. epoch.\"\"\"\n\n plt.figure()\n plt.xlabel(\"Epoch\")\n plt.ylabel(\"Root Mean Squared Error\")\n\n plt.plot(epochs, rmse, label=\"Loss\")\n plt.legend()\n plt.ylim([rmse.min()*0.97, rmse.max()])\n plt.show() \n\nprint(\"Defined the plot_the_model and plot_the_loss_curve functions.\")",
"_____no_output_____"
]
],
[
[
"## Call the model functions\n\nAn important part of machine learning is determining which [features](https://developers.google.com/machine-learning/glossary/#feature) correlate with the [label](https://developers.google.com/machine-learning/glossary/#label). For example, real-life home-value prediction models typically rely on hundreds of features and synthetic features. However, this model relies on only one feature. For now, you'll arbitrarily use `total_rooms` as that feature. \n",
"_____no_output_____"
]
],
[
[
"# The following variables are the hyperparameters.\nlearning_rate = 0.01\nepochs = 30\nbatch_size = 30\n\n# Specify the feature and the label.\nmy_feature = \"total_rooms\" # the total number of rooms on a specific city block.\nmy_label=\"median_house_value\" # the median value of a house on a specific city block.\n# That is, you're going to create a model that predicts house value based \n# solely on total_rooms. \n\n# Discard any pre-existing version of the model.\nmy_model = None\n\n# Invoke the functions.\nmy_model = build_model(learning_rate)\nweight, bias, epochs, rmse = train_model(my_model, training_df, \n my_feature, my_label,\n epochs, batch_size)\n\nprint(\"\\nThe learned weight for your model is %.4f\" % weight)\nprint(\"The learned bias for your model is %.4f\\n\" % bias )\n\nplot_the_model(weight, bias, my_feature, my_label)\nplot_the_loss_curve(epochs, rmse)",
"_____no_output_____"
]
],
[
[
"A certain amount of randomness plays into training a model. Consequently, you'll get different results each time you train the model. That said, given the dataset and the hyperparameters, the trained model will generally do a poor job describing the feature's relation to the label.",
"_____no_output_____"
],
[
"## Use the model to make predictions\n\nYou can use the trained model to make predictions. In practice, [you should make predictions on examples that are not used in training](https://developers.google.com/machine-learning/crash-course/training-and-test-sets/splitting-data). However, for this exercise, you'll just work with a subset of the same training dataset. A later Colab exercise will explore ways to make predictions on examples not used in training.\n\nFirst, run the following code to define the house prediction function:",
"_____no_output_____"
]
],
[
[
"def predict_house_values(n, feature, label):\n \"\"\"Predict house values based on a feature.\"\"\"\n\n batch = training_df[feature][10000:10000 + n]\n predicted_values = my_model.predict_on_batch(x=batch)\n\n print(\"feature label predicted\")\n print(\" value value value\")\n print(\" in thousand$ in thousand$\")\n print(\"--------------------------------------\")\n for i in range(n):\n print (\"%5.0f %6.0f %15.0f\" % (training_df[feature][10000 + i],\n training_df[label][10000 + i],\n predicted_values[i][0] ))",
"_____no_output_____"
]
],
[
[
"Now, invoke the house prediction function on 10 examples:",
"_____no_output_____"
]
],
[
[
"predict_house_values(10, my_feature, my_label)",
"_____no_output_____"
]
],
[
[
"### Task 2: Judge the predictive power of the model\n\nLook at the preceding table. How close is the predicted value to the label value? In other words, does your model accurately predict house values? ",
"_____no_output_____"
]
],
[
[
"#@title Double-click to view the answer.\n\n# Most of the predicted values differ significantly\n# from the label value, so the trained model probably \n# doesn't have much predictive power. However, the\n# first 10 examples might not be representative of \n# the rest of the examples. ",
"_____no_output_____"
]
],
[
[
"## Task 3: Try a different feature\n\nThe `total_rooms` feature had only a little predictive power. Would a different feature have greater predictive power? Try using `population` as the feature instead of `total_rooms`. \n\nNote: When you change features, you might also need to change the hyperparameters.",
"_____no_output_____"
]
],
[
[
"my_feature = \"?\" # Replace the ? with population or possibly\n # a different column name.\n\n# Experiment with the hyperparameters.\nlearning_rate = 2\nepochs = 3\nbatch_size = 120\n\n# Don't change anything below this line.\nmy_model = build_model(learning_rate)\nweight, bias, epochs, rmse = train_model(my_model, training_df, \n my_feature, my_label,\n epochs, batch_size)\nplot_the_model(weight, bias, my_feature, my_label)\nplot_the_loss_curve(epochs, rmse)\n\npredict_house_values(15, my_feature, my_label)",
"_____no_output_____"
],
[
"#@title Double-click to view a possible solution.\n\nmy_feature = \"population\" # Pick a feature other than \"total_rooms\"\n\n# Possibly, experiment with the hyperparameters.\nlearning_rate = 0.05\nepochs = 18\nbatch_size = 3\n\n# Don't change anything below.\nmy_model = build_model(learning_rate)\nweight, bias, epochs, rmse = train_model(my_model, training_df, \n my_feature, my_label,\n epochs, batch_size)\n\nplot_the_model(weight, bias, my_feature, my_label)\nplot_the_loss_curve(epochs, rmse)\n\npredict_house_values(10, my_feature, my_label)",
"_____no_output_____"
]
],
[
[
"Did `population` produce better predictions than `total_rooms`?",
"_____no_output_____"
]
],
[
[
"#@title Double-click to view the answer.\n\n# Training is not entirely deterministic, but population \n# typically converges at a slightly higher RMSE than \n# total_rooms. So, population appears to be about \n# the same or slightly worse at making predictions \n# than total_rooms.",
"_____no_output_____"
]
],
[
[
"## Task 4: Define a synthetic feature\n\nYou have determined that `total_rooms` and `population` were not useful features. That is, neither the total number of rooms in a neighborhood nor the neighborhood's population successfully predicted the median house price of that neighborhood. Perhaps though, the *ratio* of `total_rooms` to `population` might have some predictive power. That is, perhaps block density relates to median house value.\n\nTo explore this hypothesis, do the following: \n\n1. Create a [synthetic feature](https://developers.google.com/machine-learning/glossary/#synthetic_feature) that's a ratio of `total_rooms` to `population`. (If you are new to pandas DataFrames, please study the [Pandas DataFrame Ultraquick Tutorial](https://colab.research.google.com/github/google/eng-edu/blob/master/ml/cc/exercises/pandas_dataframe_ultraquick_tutorial.ipynb?utm_source=linearregressionreal-colab&utm_medium=colab&utm_campaign=colab-external&utm_content=pandas_tf2-colab&hl=en).)\n2. Tune the three hyperparameters.\n3. Determine whether this synthetic feature produces \n a lower loss value than any of the single features you \n tried earlier in this exercise.",
"_____no_output_____"
]
],
[
[
"# Define a synthetic feature named rooms_per_person\ntraining_df[\"rooms_per_person\"] = ? # write your code here.\n\n# Don't change the next line.\nmy_feature = \"rooms_per_person\"\n\n# Assign values to these three hyperparameters.\nlearning_rate = ?\nepochs = ?\nbatch_size = ?\n\n# Don't change anything below this line.\nmy_model = build_model(learning_rate)\nweight, bias, epochs, rmse = train_model(my_model, training_df,\n my_feature, my_label,\n epochs, batch_size)\n\nplot_the_loss_curve(epochs, rmse)\npredict_house_values(15, my_feature, my_label)",
"_____no_output_____"
],
[
"#@title Double-click to view a possible solution to Task 4.\n\n# Define a synthetic feature\ntraining_df[\"rooms_per_person\"] = training_df[\"total_rooms\"] / training_df[\"population\"]\nmy_feature = \"rooms_per_person\"\n\n# Tune the hyperparameters.\nlearning_rate = 0.06\nepochs = 24\nbatch_size = 30\n\n# Don't change anything below this line.\nmy_model = build_model(learning_rate)\nweight, bias, epochs, mae = train_model(my_model, training_df,\n my_feature, my_label,\n epochs, batch_size)\n\nplot_the_loss_curve(epochs, mae)\npredict_house_values(15, my_feature, my_label)\n",
"_____no_output_____"
]
],
[
[
"Based on the loss values, this synthetic feature produces a better model than the individual features you tried in Task 2 and Task 3. However, the model still isn't creating great predictions.\n",
"_____no_output_____"
],
[
"## Task 5. Find feature(s) whose raw values correlate with the label\n\nSo far, we've relied on trial-and-error to identify possible features for the model. Let's rely on statistics instead.\n\nA **correlation matrix** indicates how each attribute's raw values relate to the other attributes' raw values. Correlation values have the following meanings:\n\n * `1.0`: perfect positive correlation; that is, when one attribute rises, the other attribute rises.\n * `-1.0`: perfect negative correlation; that is, when one attribute rises, the other attribute falls. \n * `0.0`: no correlation; the two column's [are not linearly related](https://en.wikipedia.org/wiki/Correlation_and_dependence#/media/File:Correlation_examples2.svg).\n\nIn general, the higher the absolute value of a correlation value, the greater its predictive power. For example, a correlation value of -0.8 implies far more predictive power than a correlation of -0.2.\n\nThe following code cell generates the correlation matrix for attributes of the California Housing Dataset:",
"_____no_output_____"
]
],
[
[
"# Generate a correlation matrix.\ntraining_df.corr()",
"_____no_output_____"
]
],
[
[
"The correlation matrix shows nine potential features (including a synthetic\nfeature) and one label (`median_house_value`). A strong negative correlation or strong positive correlation with the label suggests a potentially good feature. \n\n**Your Task:** Determine which of the nine potential features appears to be the best candidate for a feature?",
"_____no_output_____"
]
],
[
[
"#@title Double-click here for the solution to Task 5\n\n# The `median_income` correlates 0.7 with the label \n# (median_house_value), so median_income` might be a \n# good feature. The other seven potential features\n# all have a correlation relatively close to 0. \n\n# If time permits, try median_income as the feature\n# and see whether the model improves.",
"_____no_output_____"
]
],
[
[
"Correlation matrices don't tell the entire story. In later exercises, you'll find additional ways to unlock predictive power from potential features.\n\n**Note:** Using `median_income` as a feature may raise some ethical and fairness\nissues. Towards the end of the course, we'll explore ethical and fairness issues.",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d001aa363a9fa0a5c61ece6012b44540e4d5a4c5 | 265,027 | ipynb | Jupyter Notebook | o3_so2_upb/estacion_upb_data_processing_03.ipynb | fega/arduair-calibration | 8dbcbb947fc964ab248974234c053ebde9869213 | [
"MIT"
] | null | null | null | o3_so2_upb/estacion_upb_data_processing_03.ipynb | fega/arduair-calibration | 8dbcbb947fc964ab248974234c053ebde9869213 | [
"MIT"
] | null | null | null | o3_so2_upb/estacion_upb_data_processing_03.ipynb | fega/arduair-calibration | 8dbcbb947fc964ab248974234c053ebde9869213 | [
"MIT"
] | null | null | null | 606.469108 | 65,062 | 0.935512 | [
[
[
"## Analisis de O3 y SO2 arduair vs estacion universidad pontificia bolivariana\nSe compararon los resultados generados por el equipo arduair y la estacion de calidad de aire propiedad de la universidad pontificia bolivariana seccional bucaramanga\n\nCabe resaltar que durante la ejecucion de las pruebas, el se sospechaba equipo de SO2 de la universidad pontificia, por lo cual no se pueden interpretar estos resultados como fiables ",
"_____no_output_____"
],
[
"## Library imports",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nimport datetime as dt\nimport xlrd\n%matplotlib inline\n\npd.options.mode.chained_assignment = None ",
"_____no_output_____"
]
],
[
[
"## Estudios de correlacion\nSe realizaron graficos de correlacion para el ozono y el dioxido de azufre con la estacion de referencia.\n\nTambien se comparo los datos crudos arrojados por el sensor de ozono, con las ecuaciones de calibracion propuesta por el [datasheet](https://www.terraelectronica.ru/%2Fds%2Fpdf%2FM%2Fmq131-low.pdf), obteniendose mejores resultados con los datos sin procesar.",
"_____no_output_____"
]
],
[
[
"#Arduair prototype data\ndfArd=pd.read_csv('DATA.TXT',names=['year','month','day','hour','minute','second','hum','temp','pr','l','co','so2','no2','o3','pm10','pm25','void'])\n#Dates to datetime\ndates=dfArd[['year','month','day','hour','minute','second']]\ndates['year']=dates['year'].add(2000)\ndates['minute']=dates['minute'].add(60)\ndfArd['datetime']=pd.to_datetime(dates)\n\n#agregation\ndfArdo3=dfArd[['datetime','o3']]\ndfArdso2=dfArd[['datetime','so2']]\n\n#O3 processing\nMQ131_RL= 10 #Load resistance\nMQ131_VIN = 5 #Vin\nMQ131_RO = 5 #reference resistance\ndfArdo3['rs']=((MQ131_VIN/dfArdo3['o3'])/dfArdo3['o3'])*MQ131_RL;\ndfArdo3['rs_ro'] = dfArdo3['rs']/MQ131_RO;\ndfArdo3['rs_ro_abs']=abs(dfArdo3['rs_ro'])\n\n#station data\ndfo3=pd.read_csv('o3_upb.csv')\ndfso2=pd.read_csv('so2_upb.csv')\ndfso2.tail()\ndfso2['datetime']=pd.to_datetime(dfso2['date time'])\ndfo3['datetime']=pd.to_datetime(dfo3['date time'])\n\ndfso2=dfso2[['datetime','pump_status']]\ndfo3=dfo3[['datetime','pump_status']]\n\n# bad label correction\ndfso2.columns = ['datetime', 'raw_so2']\ndfo3.columns = ['datetime', 'ozone_UPB']\n\n#grouping\ndfArdo3 =dfArdo3 .groupby(pd.Grouper(key='datetime',freq='1h',axis=1)).mean()\ndfArdso2=dfArdso2.groupby(pd.Grouper(key='datetime',freq='1h',axis=1)).mean()\ndfo3 =dfo3 .groupby(pd.Grouper(key='datetime',freq='1h',axis=1)).mean()\ndfso2 =dfso2 .groupby(pd.Grouper(key='datetime',freq='1h',axis=1)).mean()\n\ndf2=pd.concat([dfo3,dfArdo3], join='inner', axis=1).reset_index()\ndf3=pd.concat([dfso2,dfArdso2], join='inner', axis=1).reset_index()\n#Ozono calibrado\nsns.jointplot(data=df2,x='ozone_UPB',y='rs_ro', kind='reg')\n#Ozono crudo\nsns.jointplot(data=df2,x='ozone_UPB',y='o3', kind='reg')\n#SO2\nsns.jointplot(data=df3,x='raw_so2',y='so2', kind='reg')\ndfso2.head()",
"C:\\Users\\fega0\\Anaconda3\\lib\\site-packages\\statsmodels\\nonparametric\\kdetools.py:20: VisibleDeprecationWarning: using a non-integer number instead of an integer will result in an error in the future\n y = X[:m/2+1] + np.r_[0,X[m/2+1:],0]*1j\n"
]
],
[
[
"### Defino algunas funciones de ayuda",
"_____no_output_____"
]
],
[
[
"def polyfitEq(x,y):\n C= np.polyfit(x,y,1)\n m=C[0]\n b=C[1]\n return 'y = x*{} + {}'.format(m,b)\ndef calibrate(x,y):\n C= np.polyfit(x,y,1)\n m=C[0]\n b=C[1]\n return x*m+b\ndef rename_labels(obj,unit):\n obj.columns=obj.columns.map(lambda x: x.replace('2',' stc_cdmb'))\n obj.columns=obj.columns.map(lambda x: x+' '+unit)\n return obj.columns",
"_____no_output_____"
],
[
"print('')\nprint('Ozono promedio 1h, sin procesar')\nprint(polyfitEq(df2['ozone_UPB'],df2['o3']))\n#print('')\n#print('Promedio 2h')\n#print(polyfitEq(df2['pm10'],df2['pm10_dusttrack']))\nprint('')\nprint('Promedio 3h')\nprint(polyfitEq(df3['raw_so2'],df3['so2']))",
"\nOzono promedio 1h, sin procesar\ny = x*-7.386462397051218 + 735.7745124254552\n\nPromedio 3h\ny = x*3.9667587988316875 + 471.89151081632417\n"
]
],
[
[
"## Datasheets calibrados",
"_____no_output_____"
]
],
[
[
"df2['o3']=calibrate(df2['o3'],df2['ozone_UPB'])\ndf2.plot(figsize=[15,5])\n\ndf3['so2']=calibrate(df3['so2'],df3['raw_so2'])\ndf3.plot(figsize=[15,5])\n\n",
"_____no_output_____"
],
[
"df2.head()\ndf2.columns = ['datetime', 'Ozono estación UPB [ppb]','Ozono prototipo [ppb]','rs','rs_ro','rs_ro_abs']\nsns.jointplot(data=df2,x='Ozono prototipo [ppb]',y='Ozono estación UPB [ppb]', kind='reg',stat_func=None)",
"C:\\Users\\fega0\\Anaconda3\\lib\\site-packages\\statsmodels\\nonparametric\\kdetools.py:20: VisibleDeprecationWarning: using a non-integer number instead of an integer will result in an error in the future\n y = X[:m/2+1] + np.r_[0,X[m/2+1:],0]*1j\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d001ace14ccebd1da740b49bc211312a9f784676 | 3,445 | ipynb | Jupyter Notebook | examples/discovery v1 configuration tasks .ipynb | SeptBlast/python-sdk | 8ba86b8abbff7cd020303b877d730130696ea21d | [
"Apache-2.0"
] | 4 | 2019-03-19T05:07:32.000Z | 2021-08-12T13:11:30.000Z | pythonQueryServices/pythonWatson/pythonSDK/python-sdk-master/examples/discovery v1 configuration tasks .ipynb | xliuhw/NLU-Evaluation-Scripts | 356711b59f347532d0290f070ff9aad5af7ed02e | [
"MIT"
] | null | null | null | pythonQueryServices/pythonWatson/pythonSDK/python-sdk-master/examples/discovery v1 configuration tasks .ipynb | xliuhw/NLU-Evaluation-Scripts | 356711b59f347532d0290f070ff9aad5af7ed02e | [
"MIT"
] | null | null | null | 23.59589 | 125 | 0.584615 | [
[
[
"import os,sys\nsys.path.append(os.path.join(os.getcwd(),'..'))\nimport watson_developer_cloud\n\nDISCOVERY_USERNAME='CHANGE_ME'\nDISCOVERY_PASSWORD='CHANGE_ME'\nENVIRONMENT_NAME='CHANGE_ME' # this is the 'name' field of your environment\nCONFIGURATION_NAME='CHANGE_ME' # this is the 'name' field of your cofiguration",
"_____no_output_____"
],
[
"discovery = watson_developer_cloud.DiscoveryV1(\n '2016-12-15',\n username=DISCOVERY_USERNAME,\n password=DISCOVERY_PASSWORD)",
"_____no_output_____"
],
[
"environments = discovery.get_environments()\nprint(environments)",
"_____no_output_____"
],
[
"target_environment = [x for x in environments['environments'] if x['name'] == ENVIRONMENT_NAME]\ntarget_environment_id = target_environment[0]['environment_id']\nprint(target_environment_id)",
"_____no_output_____"
],
[
"configs = discovery.list_configurations(environment_id=target_environment_id)\nprint(configs)",
"_____no_output_____"
],
[
"target_config = [x for x in configs['configurations'] if x['name'] == CONFIGURATION_NAME]\ntarget_config_id = target_config[0]['configuration_id']\nprint(target_config_id)",
"_____no_output_____"
],
[
"config_data = discovery.get_configuration(environment_id=target_environment_id,\n configuration_id=target_config_id)\nprint(config_data)",
"_____no_output_____"
],
[
"config_data['name'] = 'Changed Name for Example'\nres = discovery.create_configuration(environment_id=target_environment_id, config_data=config_data)\nprint(res)",
"_____no_output_____"
],
[
"res = discovery.delete_configuration(environment_id=target_environment_id, configuration_id=res['configuration_id'])\nprint(res)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d001c5012581478f36d371e7ce2c756d6ab6ad7e | 1,035,343 | ipynb | Jupyter Notebook | datasets/terraclimate/terraclimate-example.ipynb | ianthomas23/PlanetaryComputerExamples | 490f353c95ef2de48374c45818e35aef90cf424a | [
"MIT"
] | 112 | 2021-04-15T19:58:38.000Z | 2022-03-30T18:00:31.000Z | datasets/terraclimate/terraclimate-example.ipynb | ruiduobao/PlanetaryComputerExamples | 2296dfbd4824f840954b07dcb85ce1b8c6f4e628 | [
"MIT"
] | 61 | 2021-04-19T13:55:30.000Z | 2022-03-31T23:52:48.000Z | datasets/terraclimate/terraclimate-example.ipynb | ruiduobao/PlanetaryComputerExamples | 2296dfbd4824f840954b07dcb85ce1b8c6f4e628 | [
"MIT"
] | 54 | 2021-04-19T20:37:46.000Z | 2022-03-28T19:47:09.000Z | 334.413114 | 348,644 | 0.867455 | [
[
[
"## Accessing TerraClimate data with the Planetary Computer STAC API\n\n[TerraClimate](http://www.climatologylab.org/terraclimate.html) is a dataset of monthly climate and climatic water balance for global terrestrial surfaces from 1958-2019. These data provide important inputs for ecological and hydrological studies at global scales that require high spatial resolution and time-varying data. All data have monthly temporal resolution and a ~4-km (1/24th degree) spatial resolution. The data cover the period from 1958-2019.\n\nThis example will show you how temperature has increased over the past 60 years across the globe.\n\n### Environment setup",
"_____no_output_____"
]
],
[
[
"import warnings\n\nwarnings.filterwarnings(\"ignore\", \"invalid value\", RuntimeWarning)",
"_____no_output_____"
]
],
[
[
"### Data access\n\nhttps://planetarycomputer.microsoft.com/api/stac/v1/collections/terraclimate is a STAC Collection with links to all the metadata about this dataset. We'll load it with [PySTAC](https://pystac.readthedocs.io/en/latest/).",
"_____no_output_____"
]
],
[
[
"import pystac\n\nurl = \"https://planetarycomputer.microsoft.com/api/stac/v1/collections/terraclimate\"\ncollection = pystac.read_file(url)\ncollection",
"_____no_output_____"
]
],
[
[
"The collection contains assets, which are links to the root of a Zarr store, which can be opened with xarray.",
"_____no_output_____"
]
],
[
[
"asset = collection.assets[\"zarr-https\"]\nasset",
"_____no_output_____"
],
[
"import fsspec\nimport xarray as xr\n\nstore = fsspec.get_mapper(asset.href)\nds = xr.open_zarr(store, **asset.extra_fields[\"xarray:open_kwargs\"])\nds",
"_____no_output_____"
]
],
[
[
"We'll process the data in parallel using [Dask](https://dask.org).",
"_____no_output_____"
]
],
[
[
"from dask_gateway import GatewayCluster\n\ncluster = GatewayCluster()\ncluster.scale(16)\nclient = cluster.get_client()\nprint(cluster.dashboard_link)",
"https://pcc-staging.westeurope.cloudapp.azure.com/compute/services/dask-gateway/clusters/staging.5cae9b2b4c7d4f7fa37c5a4ac1e8112d/status\n"
]
],
[
[
"The link printed out above can be opened in a new tab or the [Dask labextension](https://github.com/dask/dask-labextension). See [Scale with Dask](https://planetarycomputer.microsoft.com/docs/quickstarts/scale-with-dask/) for more on using Dask, and how to access the Dashboard.\n\n### Analyze and plot global temperature\n\nWe can quickly plot a map of one of the variables. In this case, we are downsampling (coarsening) the dataset for easier plotting.",
"_____no_output_____"
]
],
[
[
"import cartopy.crs as ccrs\nimport matplotlib.pyplot as plt\n\n\naverage_max_temp = ds.isel(time=-1)[\"tmax\"].coarsen(lat=8, lon=8).mean().load()\n\nfig, ax = plt.subplots(figsize=(20, 10), subplot_kw=dict(projection=ccrs.Robinson()))\n\naverage_max_temp.plot(ax=ax, transform=ccrs.PlateCarree())\nax.coastlines();",
"_____no_output_____"
]
],
[
[
"Let's see how temperature has changed over the observational record, when averaged across the entire domain. Since we'll do some other calculations below we'll also add `.load()` to execute the command instead of specifying it lazily. Note that there are some data quality issues before 1965 so we'll start our analysis there.",
"_____no_output_____"
]
],
[
[
"temperature = (\n ds[\"tmax\"].sel(time=slice(\"1965\", None)).mean(dim=[\"lat\", \"lon\"]).persist()\n)",
"_____no_output_____"
],
[
"temperature.plot(figsize=(12, 6));",
"_____no_output_____"
]
],
[
[
"With all the seasonal fluctuations (from summer and winter) though, it can be hard to see any obvious trends. So let's try grouping by year and plotting that timeseries.",
"_____no_output_____"
]
],
[
[
"temperature.groupby(\"time.year\").mean().plot(figsize=(12, 6));",
"_____no_output_____"
]
],
[
[
"Now the increase in temperature is obvious, even when averaged across the entire domain.\n\nNow, let's see how those changes are different in different parts of the world. And let's focus just on summer months in the northern hemisphere, when it's hottest. Let's take a climatological slice at the beginning of the period and the same at the end of the period, calculate the difference, and map it to see how different parts of the world have changed differently.\n\nFirst we'll just grab the summer months.",
"_____no_output_____"
]
],
[
[
"%%time\nimport dask\n\nsummer_months = [6, 7, 8]\nsummer = ds.tmax.where(ds.time.dt.month.isin(summer_months), drop=True)\n\nearly_period = slice(\"1958-01-01\", \"1988-12-31\")\nlate_period = slice(\"1988-01-01\", \"2018-12-31\")\n\nearly, late = dask.compute(\n summer.sel(time=early_period).mean(dim=\"time\"),\n summer.sel(time=late_period).mean(dim=\"time\"),\n)\nincrease = (late - early).coarsen(lat=8, lon=8).mean()",
"CPU times: user 1.35 s, sys: 365 ms, total: 1.71 s\nWall time: 32.8 s\n"
],
[
"fig, ax = plt.subplots(figsize=(20, 10), subplot_kw=dict(projection=ccrs.Robinson()))\n\nincrease.plot(ax=ax, transform=ccrs.PlateCarree(), robust=True)\nax.coastlines();",
"_____no_output_____"
]
],
[
[
"This shows us that changes in summer temperature haven't been felt equally around the globe. Note the enhanced warming in the polar regions, a phenomenon known as \"Arctic amplification\".",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d001cb3360293f2ae9e91ec462e9e3bfafdf84f4 | 12,880 | ipynb | Jupyter Notebook | notebooks/automl-classification-Force-text-dnn.ipynb | konabuta/AutoML-Pipeline | 4cb9a3ccc15d482f0b4d1fcacd53ff81f28b14be | [
"MIT"
] | 2 | 2020-10-10T16:47:37.000Z | 2020-10-14T02:49:58.000Z | notebooks/automl-classification-Force-text-dnn.ipynb | konabuta/AutoML-Pipeline | 4cb9a3ccc15d482f0b4d1fcacd53ff81f28b14be | [
"MIT"
] | null | null | null | notebooks/automl-classification-Force-text-dnn.ipynb | konabuta/AutoML-Pipeline | 4cb9a3ccc15d482f0b4d1fcacd53ff81f28b14be | [
"MIT"
] | null | null | null | 24.769231 | 228 | 0.564053 | [
[
[
"Copyright (c) Microsoft Corporation. All rights reserved.\n\nLicensed under the MIT License.",
"_____no_output_____"
],
[
"# Automated Machine Learning\n_**ディープラーンニングを利用したテキスト分類**_\n\n## Contents\n1. [事前準備](#1.-事前準備)\n1. [自動機械学習 Automated Machine Learning](2.-自動機械学習-Automated-Machine-Learning)\n1. [結果の確認](#3.-結果の確認)",
"_____no_output_____"
],
[
"## 1. 事前準備\n\n本デモンストレーションでは、AutoML の深層学習の機能を用いてテキストデータの分類モデルを構築します。 \nAutoML には Deep Neural Network が含まれており、テキストデータから **Embedding** を作成することができます。GPU サーバを利用することで **BERT** が利用されます。\n\n深層学習の機能を利用するためには Azure Machine Learning の Enterprise Edition が必要になります。詳細は[こちら](https://docs.microsoft.com/en-us/azure/machine-learning/concept-editions#automated-training-capabilities-automl)をご確認ください。",
"_____no_output_____"
],
[
"## 1.1 Python SDK のインポート",
"_____no_output_____"
],
[
"Azure Machine Learning の Python SDK などをインポートします。",
"_____no_output_____"
]
],
[
[
"import logging\nimport os\nimport shutil\n\nimport pandas as pd\n\nimport azureml.core\nfrom azureml.core.experiment import Experiment\nfrom azureml.core.workspace import Workspace\nfrom azureml.core.dataset import Dataset\nfrom azureml.core.compute import AmlCompute\nfrom azureml.core.compute import ComputeTarget\nfrom azureml.core.run import Run\nfrom azureml.widgets import RunDetails\nfrom azureml.core.model import Model \nfrom azureml.train.automl import AutoMLConfig\nfrom sklearn.datasets import fetch_20newsgroups",
"_____no_output_____"
],
[
"from azureml.automl.core.featurization import FeaturizationConfig",
"_____no_output_____"
]
],
[
[
"Azure ML Python SDK のバージョンが 1.8.0 以上になっていることを確認します。",
"_____no_output_____"
]
],
[
[
"print(\"You are currently using version\", azureml.core.VERSION, \"of the Azure ML SDK\")",
"_____no_output_____"
]
],
[
[
"## 1.2 Azure ML Workspace との接続",
"_____no_output_____"
]
],
[
[
"ws = Workspace.from_config()",
"_____no_output_____"
],
[
"\n# 実験名の指定\nexperiment_name = 'livedoor-news-classification-BERT'\n\nexperiment = Experiment(ws, experiment_name)\n\noutput = {}\n#output['Subscription ID'] = ws.subscription_id\noutput['Workspace Name'] = ws.name\noutput['Resource Group'] = ws.resource_group\noutput['Location'] = ws.location\noutput['Experiment Name'] = experiment.name\npd.set_option('display.max_colwidth', -1)\noutputDf = pd.DataFrame(data = output, index = [''])\noutputDf.T",
"_____no_output_____"
]
],
[
[
"## 1.3 計算環境の準備\n\nBERT を利用するための GPU の `Compute Cluster` を準備します。",
"_____no_output_____"
]
],
[
[
"from azureml.core.compute import ComputeTarget, AmlCompute\nfrom azureml.core.compute_target import ComputeTargetException\n\n# Compute Cluster の名称\namlcompute_cluster_name = \"gpucluster\"\n\n# クラスターの存在確認\ntry:\n compute_target = ComputeTarget(workspace=ws, name=amlcompute_cluster_name)\n \nexcept ComputeTargetException:\n print('指定された名称のクラスターが見つからないので新規に作成します.')\n compute_config = AmlCompute.provisioning_configuration(vm_size = \"STANDARD_NC6_V3\",\n max_nodes = 4)\n compute_target = ComputeTarget.create(ws, amlcompute_cluster_name, compute_config)\n\ncompute_target.wait_for_completion(show_output=True)",
"_____no_output_____"
]
],
[
[
"## 1.4 学習データの準備\n今回は [livedoor New](https://www.rondhuit.com/download/ldcc-20140209.tar.gz) を学習データとして利用します。ニュースのカテゴリー分類のモデルを構築します。 ",
"_____no_output_____"
]
],
[
[
"target_column_name = 'label' # カテゴリーの列 \nfeature_column_name = 'text' # ニュース記事の列",
"_____no_output_____"
],
[
"train_dataset = Dataset.get_by_name(ws, \"livedoor\").keep_columns([\"text\",\"label\"])\ntrain_dataset.take(5).to_pandas_dataframe()",
"_____no_output_____"
]
],
[
[
"# 2. 自動機械学習 Automated Machine Learning\n## 2.1 設定と制約条件",
"_____no_output_____"
],
[
"自動機械学習 Automated Machine Learning の設定と学習を行っていきます。",
"_____no_output_____"
]
],
[
[
"from azureml.automl.core.featurization import FeaturizationConfig\nfeaturization_config = FeaturizationConfig()\n# テキストデータの言語を指定します。日本語の場合は \"jpn\" と指定します。\nfeaturization_config = FeaturizationConfig(dataset_language=\"jpn\") # 英語の場合は下記をコメントアウトしてください。",
"_____no_output_____"
],
[
"# 明示的に `text` の列がテキストデータであると指定します。\nfeaturization_config.add_column_purpose('text', 'Text')\n#featurization_config.blocked_transformers = ['TfIdf','CountVectorizer'] # BERT のみを利用したい場合はコメントアウトを外します",
"_____no_output_____"
],
[
"# 自動機械学習の設定\nautoml_settings = {\n \"experiment_timeout_hours\" : 2, # 学習時間 (hour)\n \"primary_metric\": 'accuracy', # 評価指標\n \"max_concurrent_iterations\": 4, # 計算環境の最大並列数 \n \"max_cores_per_iteration\": -1,\n \"enable_dnn\": True, # 深層学習を有効\n \"enable_early_stopping\": False,\n \"validation_size\": 0.2,\n \"verbosity\": logging.INFO,\n \"force_text_dnn\": True,\n #\"n_cross_validations\": 5,\n}\n\nautoml_config = AutoMLConfig(task = 'classification', \n debug_log = 'automl_errors.log',\n compute_target=compute_target,\n training_data=train_dataset,\n label_column_name=target_column_name,\n featurization=featurization_config,\n **automl_settings\n )",
"_____no_output_____"
]
],
[
[
"## 2.2 モデル学習",
"_____no_output_____"
],
[
"自動機械学習 Automated Machine Learning によるモデル学習を開始します。",
"_____no_output_____"
]
],
[
[
"automl_run = experiment.submit(automl_config, show_output=False)",
"_____no_output_____"
],
[
"# run_id を出力\nautoml_run.id",
"_____no_output_____"
],
[
"# Azure Machine Learning studio の URL を出力\nautoml_run",
"_____no_output_____"
],
[
"# # 途中でセッションが切れた場合の対処\n# from azureml.train.automl.run import AutoMLRun\n# ws = Workspace.from_config()\n# experiment = ws.experiments['livedoor-news-classification-BERT'] \n# run_id = \"AutoML_e69a63ae-ef52-4783-9a9f-527d69d7cc9d\"\n# automl_run = AutoMLRun(experiment, run_id = run_id)\n# automl_run\n",
"_____no_output_____"
]
],
[
[
"## 2.3 モデルの登録",
"_____no_output_____"
]
],
[
[
"# 一番精度が高いモデルを抽出\nbest_run, fitted_model = automl_run.get_output()",
"_____no_output_____"
],
[
"# モデルファイル(.pkl) のダウンロード\nmodel_dir = '../model'\nbest_run.download_file('outputs/model.pkl', model_dir + '/model.pkl')",
"_____no_output_____"
],
[
"# Azure ML へモデル登録\nmodel_name = 'livedoor-model'\nmodel = Model.register(model_path = model_dir + '/model.pkl',\n model_name = model_name,\n tags=None,\n workspace=ws)",
"_____no_output_____"
]
],
[
[
"# 3. テストデータに対する予測値の出力",
"_____no_output_____"
]
],
[
[
"from sklearn.externals import joblib\ntrained_model = joblib.load(model_dir + '/model.pkl')",
"_____no_output_____"
],
[
"trained_model",
"_____no_output_____"
],
[
"test_dataset = Dataset.get_by_name(ws, \"livedoor\").keep_columns([\"text\"])\npredicted = trained_model.predict_proba(test_dataset.to_pandas_dataframe())",
"_____no_output_____"
]
],
[
[
"# 4. モデルの解釈\n一番精度が良かったチャンピョンモデルを選択し、モデルの解釈をしていきます。 \nモデルに含まれるライブラリを予め Python 環境にインストールする必要があります。[automl_env.yml](https://github.com/Azure/MachineLearningNotebooks/blob/master/how-to-use-azureml/automated-machine-learning/automl_env.yml)を用いて、conda の仮想環境に必要なパッケージをインストールしてください。",
"_____no_output_____"
]
],
[
[
"# 特徴量エンジニアリング後の変数名の確認\nfitted_model.named_steps['datatransformer'].get_json_strs_for_engineered_feature_names()\n#fitted_model.named_steps['datatransformer']. get_engineered_feature_names ()",
"_____no_output_____"
],
[
"# 特徴エンジニアリングのプロセスの可視化\ntext_transformations_used = []\nfor column_group in fitted_model.named_steps['datatransformer'].get_featurization_summary():\n text_transformations_used.extend(column_group['Transformations'])\ntext_transformations_used",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d001dafa97a6c07fba51044eadcdf8d3aad367a9 | 74,599 | ipynb | Jupyter Notebook | notebooks/colab/automl-baseline/process_autogluon_results.ipynb | pszemraj/ml4hc-s22-project01 | 219d5b7bb1de05aba300b52ec6527c4fad5cca15 | [
"Apache-2.0"
] | null | null | null | notebooks/colab/automl-baseline/process_autogluon_results.ipynb | pszemraj/ml4hc-s22-project01 | 219d5b7bb1de05aba300b52ec6527c4fad5cca15 | [
"Apache-2.0"
] | null | null | null | notebooks/colab/automl-baseline/process_autogluon_results.ipynb | pszemraj/ml4hc-s22-project01 | 219d5b7bb1de05aba300b52ec6527c4fad5cca15 | [
"Apache-2.0"
] | null | null | null | 37.187936 | 509 | 0.357833 | [
[
[
"<a href=\"https://colab.research.google.com/github/pszemraj/ml4hc-s22-project01/blob/autogluon-results/notebooks/colab/automl-baseline/process_autogluon_results.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"#process_autogluon_results\n\n- cleans up the dataframes a bit for the report\n",
"_____no_output_____"
],
[
"# setup",
"_____no_output_____"
]
],
[
[
"#@markdown add auto-Colab formatting with `IPython.display`\nfrom IPython.display import HTML, display\n# colab formatting\ndef set_css():\n display(\n HTML(\n \"\"\"\n <style>\n pre {\n white-space: pre-wrap;\n }\n </style>\n \"\"\"\n )\n )\n\nget_ipython().events.register(\"pre_run_cell\", set_css)",
"_____no_output_____"
],
[
"!nvidia-smi",
"_____no_output_____"
],
[
"!pip install -U plotly orca kaleido -q\nimport plotly.express as px",
"_____no_output_____"
],
[
"import numpy as np \nimport pandas as pd\nfrom pathlib import Path\nimport os",
"_____no_output_____"
],
[
"#@title mount drive\nfrom google.colab import drive\n\ndrive_base_str = '/content/drive'\ndrive.mount(drive_base_str)\n",
"_____no_output_____"
],
[
"#@markdown determine root\nimport os\nfrom pathlib import Path\npeter_base = Path('/content/drive/MyDrive/ETHZ-2022-S/ML-healthcare-projects/project1/gluon-autoML/')\n\nif peter_base.exists() and peter_base.is_dir():\n path = str(peter_base.resolve())\nelse:\n # original\n path = '/content/drive/MyDrive/ETH/'\n\nprint(f\"base drive dir is:\\n{path}\")",
"_____no_output_____"
]
],
[
[
"## define folder for outputs",
"_____no_output_____"
]
],
[
[
"_out_dir_name = \"Formatted-results-report\" #@param {type:\"string\"}\n\noutput_path = os.path.join(path, _out_dir_name)\nos.makedirs(output_path, exist_ok=True)\nprint(f\"notebook outputs will be stored in:\\n{output_path}\")",
"_____no_output_____"
],
[
"_out = Path(output_path)\n_src = Path(path)",
"_____no_output_____"
]
],
[
[
"##load data",
"_____no_output_____"
],
[
"### MIT",
"_____no_output_____"
]
],
[
[
"data_dir = _src / \"final-results\"\n\ncsv_files = {f.stem:f for f in data_dir.iterdir() if f.is_file() and f.suffix=='.csv'}\n\nprint(csv_files)",
"_____no_output_____"
],
[
"mit_ag = pd.read_csv(csv_files['mitbih_autogluon_results'])\nmit_ag.info()\n",
"_____no_output_____"
],
[
"mit_ag.sort_values(by='score_val', ascending=False, inplace=True)\nmit_ag.head()",
"_____no_output_____"
],
[
"orig_cols = list(mit_ag.columns)\nnew_cols = []\nfor i, col in enumerate(orig_cols):\n col = col.lower()\n if 'unnamed' in col:\n new_cols.append(f\"delete_me_{i}\")\n continue\n col = col.replace('score', 'accuracy')\n new_cols.append(col)\n\nmit_ag.columns = new_cols",
"_____no_output_____"
],
[
"mit_ag.columns",
"_____no_output_____"
],
[
"try:\n del mit_ag['delete_me_0']\nexcept Exception as e:\n print(f'skipping delete - {e}')",
"_____no_output_____"
],
[
"mit_ag.reset_index(drop=True, inplace=True)\nmit_ag.head()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"#### save mit-gluon-reformat",
"_____no_output_____"
]
],
[
[
"mit_ag.to_csv(_out / \"MITBIH_autogluon_baseline_results_Accuracy.csv\", index=False)",
"_____no_output_____"
]
],
[
[
"## PTB reformat",
"_____no_output_____"
]
],
[
[
"ptb_ag = pd.read_csv(csv_files['ptbdb_autogluon_results']).convert_dtypes()\nptb_ag.info()",
"_____no_output_____"
],
[
"ptb_ag.sort_values(by='score_val', ascending=False, inplace=True)\nptb_ag.head()",
"_____no_output_____"
],
[
"orig_cols = list(ptb_ag.columns)\nnew_cols = []\nfor i, col in enumerate(orig_cols):\n col = col.lower()\n if 'unnamed' in col:\n new_cols.append(f\"delete_me_{i}\")\n continue\n col = col.replace('score', 'roc_auc')\n new_cols.append(col)\n\nptb_ag.columns = new_cols\nprint(f'the columns for the ptb results are now:\\n{ptb_ag.columns}')",
"_____no_output_____"
],
[
"try:\n del ptb_ag['delete_me_0']\nexcept Exception as e:\n print(f'skipping delete - {e}')\n\n\nptb_ag.reset_index(drop=True, inplace=True)\nptb_ag.head()",
"_____no_output_____"
],
[
"ptb_ag.to_csv(_out / \"PTBDB_autogluon_baseline_results_ROCAUC.csv\", index=False)\n",
"_____no_output_____"
],
[
"print(f'results are in {_out.resolve()}')",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d001ded4ca679855d7525d0b9b8f0582b79578c0 | 16,653 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive/02_generalization/repeatable_splitting.ipynb | AmirQureshi/code-to-run- | bc8e5ee5b55c0408b7436d0f866b3b7e79164daf | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive/02_generalization/repeatable_splitting.ipynb | AmirQureshi/code-to-run- | bc8e5ee5b55c0408b7436d0f866b3b7e79164daf | [
"Apache-2.0"
] | null | null | null | courses/machine_learning/deepdive/02_generalization/repeatable_splitting.ipynb | AmirQureshi/code-to-run- | bc8e5ee5b55c0408b7436d0f866b3b7e79164daf | [
"Apache-2.0"
] | null | null | null | 35.583333 | 581 | 0.525791 | [
[
[
"<h1> Repeatable splitting </h1>\n\nIn this notebook, we will explore the impact of different ways of creating machine learning datasets.\n\n<p>\n\nRepeatability is important in machine learning. If you do the same thing now and 5 minutes from now and get different answers, then it makes experimentation is difficult. In other words, you will find it difficult to gauge whether a change you made has resulted in an improvement or not.",
"_____no_output_____"
]
],
[
[
"import google.datalab.bigquery as bq",
"_____no_output_____"
]
],
[
[
"<h3> Create a simple machine learning model </h3>\n\nThe dataset that we will use is <a href=\"https://bigquery.cloud.google.com/table/bigquery-samples:airline_ontime_data.flights\">a BigQuery public dataset</a> of airline arrival data. Click on the link, and look at the column names. Switch to the Details tab to verify that the number of records is 70 million, and then switch to the Preview tab to look at a few rows.\n<p>\nWe want to predict the arrival delay of an airline based on the departure delay. The model that we will use is a zero-bias linear model:\n$$ delay_{arrival} = \\alpha * delay_{departure} $$\n<p>\nTo train the model is to estimate a good value for $\\alpha$. \n<p>\nOne approach to estimate alpha is to use this formula:\n$$ \\alpha = \\frac{\\sum delay_{departure} delay_{arrival} }{ \\sum delay_{departure}^2 } $$\nBecause we'd like to capture the idea that this relationship is different for flights from New York to Los Angeles vs. flights from Austin to Indianapolis (shorter flight, less busy airports), we'd compute a different $alpha$ for each airport-pair. For simplicity, we'll do this model only for flights between Denver and Los Angeles.",
"_____no_output_____"
],
[
"<h2> Naive random split (not repeatable) </h2>",
"_____no_output_____"
]
],
[
[
"compute_alpha = \"\"\"\n#standardSQL\nSELECT \n SAFE_DIVIDE(SUM(arrival_delay * departure_delay), SUM(departure_delay * departure_delay)) AS alpha\nFROM\n(\n SELECT RAND() AS splitfield,\n arrival_delay,\n departure_delay\nFROM\n `bigquery-samples.airline_ontime_data.flights`\nWHERE\n departure_airport = 'DEN' AND arrival_airport = 'LAX'\n)\nWHERE\n splitfield < 0.8\n\"\"\"",
"_____no_output_____"
],
[
"results = bq.Query(compute_alpha).execute().result().to_dataframe()\nalpha = results['alpha'][0]\nprint alpha",
"0.975701430281\n"
]
],
[
[
"<h3> What is wrong with calculating RMSE on the training and test data as follows? </h3>",
"_____no_output_____"
]
],
[
[
"compute_rmse = \"\"\"\n#standardSQL\nSELECT\n dataset,\n SQRT(AVG((arrival_delay - ALPHA * departure_delay)*(arrival_delay - ALPHA * departure_delay))) AS rmse,\n COUNT(arrival_delay) AS num_flights\nFROM (\n SELECT\n IF (RAND() < 0.8, 'train', 'eval') AS dataset,\n arrival_delay,\n departure_delay\n FROM\n `bigquery-samples.airline_ontime_data.flights`\n WHERE\n departure_airport = 'DEN'\n AND arrival_airport = 'LAX' )\nGROUP BY\n dataset\n\"\"\"\nbq.Query(compute_rmse.replace('ALPHA', str(alpha))).execute().result()",
"_____no_output_____"
]
],
[
[
"Hint:\n* Are you really getting the same training data in the compute_rmse query as in the compute_alpha query?\n* Do you get the same answers each time you rerun the compute_alpha and compute_rmse blocks?",
"_____no_output_____"
],
[
"<h3> How do we correctly train and evaluate? </h3>\n<br/>\nHere's the right way to compute the RMSE using the actual training and held-out (evaluation) data. Note how much harder this feels.\n\nAlthough the calculations are now correct, the experiment is still not repeatable.\n\nTry running it several times; do you get the same answer?",
"_____no_output_____"
]
],
[
[
"train_and_eval_rand = \"\"\"\n#standardSQL\nWITH\n alldata AS (\n SELECT\n IF (RAND() < 0.8,\n 'train',\n 'eval') AS dataset,\n arrival_delay,\n departure_delay\n FROM\n `bigquery-samples.airline_ontime_data.flights`\n WHERE\n departure_airport = 'DEN'\n AND arrival_airport = 'LAX' ),\n training AS (\n SELECT\n SAFE_DIVIDE( SUM(arrival_delay * departure_delay) , SUM(departure_delay * departure_delay)) AS alpha\n FROM\n alldata\n WHERE\n dataset = 'train' )\nSELECT\n MAX(alpha) AS alpha,\n dataset,\n SQRT(AVG((arrival_delay - alpha * departure_delay)*(arrival_delay - alpha * departure_delay))) AS rmse,\n COUNT(arrival_delay) AS num_flights\nFROM\n alldata,\n training\nGROUP BY\n dataset\n\"\"\"",
"_____no_output_____"
],
[
"bq.Query(train_and_eval_rand).execute().result()",
"_____no_output_____"
]
],
[
[
"<h2> Using HASH of date to split the data </h2>\n\nLet's split by date and train.",
"_____no_output_____"
]
],
[
[
"compute_alpha = \"\"\"\n#standardSQL\nSELECT \n SAFE_DIVIDE(SUM(arrival_delay * departure_delay), SUM(departure_delay * departure_delay)) AS alpha\nFROM\n `bigquery-samples.airline_ontime_data.flights`\nWHERE\n departure_airport = 'DEN' AND arrival_airport = 'LAX'\n AND MOD(ABS(FARM_FINGERPRINT(date)), 10) < 8\n\"\"\"\nresults = bq.Query(compute_alpha).execute().result().to_dataframe()\nalpha = results['alpha'][0]\nprint alpha",
"0.975803914362\n"
]
],
[
[
"We can now use the alpha to compute RMSE. Because the alpha value is repeatable, we don't need to worry that the alpha in the compute_rmse will be different from the alpha computed in the compute_alpha.",
"_____no_output_____"
]
],
[
[
"compute_rmse = \"\"\"\n#standardSQL\nSELECT\n IF(MOD(ABS(FARM_FINGERPRINT(date)), 10) < 8, 'train', 'eval') AS dataset,\n SQRT(AVG((arrival_delay - ALPHA * departure_delay)*(arrival_delay - ALPHA * departure_delay))) AS rmse,\n COUNT(arrival_delay) AS num_flights\nFROM\n `bigquery-samples.airline_ontime_data.flights`\nWHERE\n departure_airport = 'DEN'\n AND arrival_airport = 'LAX'\nGROUP BY\n dataset\n\"\"\"\nprint bq.Query(compute_rmse.replace('ALPHA', str(alpha))).execute().result().to_dataframe().head()",
" dataset rmse num_flights\n0 eval 12.764685 15671\n1 train 13.160712 64018\n"
]
],
[
[
"Note also that the RMSE on the evaluation dataset more from the RMSE on the training dataset when we do the split correctly. This should be expected; in the RAND() case, there was leakage between training and evaluation datasets, because there is high correlation between flights on the same day.\n<p>\nThis is one of the biggest dangers with doing machine learning splits the wrong way -- <b> you will develop a false sense of confidence in how good your model is! </b>",
"_____no_output_____"
],
[
"Copyright 2017 Google Inc.\nLicensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at\nhttp://www.apache.org/licenses/LICENSE-2.0\nUnless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d001e1dab2b894cab4d4d2c83c742e1185d5b2cb | 10,010 | ipynb | Jupyter Notebook | examples/notebook/contrib/steel.ipynb | MaximilianAzendorf/wasm-or-tools | f16c3efc13ad5d41c7a65338434ea88ed908c398 | [
"Apache-2.0"
] | null | null | null | examples/notebook/contrib/steel.ipynb | MaximilianAzendorf/wasm-or-tools | f16c3efc13ad5d41c7a65338434ea88ed908c398 | [
"Apache-2.0"
] | null | null | null | examples/notebook/contrib/steel.ipynb | MaximilianAzendorf/wasm-or-tools | f16c3efc13ad5d41c7a65338434ea88ed908c398 | [
"Apache-2.0"
] | null | null | null | 38.206107 | 243 | 0.535964 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d001e40a54bddb838a8e4e457fafea447a6e0f3a | 14,298 | ipynb | Jupyter Notebook | _drafts/linear-optimization/.ipynb_checkpoints/Linear Optimization-checkpoint.ipynb | evjrob/everettsprojects.com | 95b22907bd9f8b4aa2e3df510c2c263267a3775e | [
"MIT"
] | 2 | 2021-02-05T08:40:37.000Z | 2021-03-30T09:19:23.000Z | _drafts/linear-optimization/.ipynb_checkpoints/Linear Optimization-checkpoint.ipynb | evjrob/everettsprojects.com | 95b22907bd9f8b4aa2e3df510c2c263267a3775e | [
"MIT"
] | 9 | 2020-08-08T22:27:14.000Z | 2022-03-12T00:59:27.000Z | _drafts/linear-optimization/.ipynb_checkpoints/Linear Optimization-checkpoint.ipynb | evjrob/everettsprojects.com | 95b22907bd9f8b4aa2e3df510c2c263267a3775e | [
"MIT"
] | null | null | null | 164.344828 | 12,520 | 0.889145 | [
[
[
"import networkx as nx\nimport matplotlib.pyplot as plt\n\nG=nx.Graph()\nG.add_node(\"CPF\")\n\nG.add_nodes_from([\"Pad1\",\n \"Pad2\",\n \"Pad3\",\n \"Pad4\",\n \"Pad5\",\n \"Pad6\"])\n\nG.add_edges_from([(\"CPF\",\"Pad1\"),\n (\"CPF\",\"Pad2\"),\n (\"Pad1\",\"Pad3\"),\n (\"Pad1\",\"Pad4\"),\n (\"Pad2\",\"Pad5\"),\n (\"Pad2\",\"Pad6\")])\nnx.draw(G)\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d001efd8179f1c0b37fcf3b8a9cfb25aae925b34 | 14,560 | ipynb | Jupyter Notebook | data/read_data.ipynb | sannatti/softcifar | 6d93cc6732b8487a4369960dcfaa6cc8f1f65164 | [
"MIT"
] | null | null | null | data/read_data.ipynb | sannatti/softcifar | 6d93cc6732b8487a4369960dcfaa6cc8f1f65164 | [
"MIT"
] | null | null | null | data/read_data.ipynb | sannatti/softcifar | 6d93cc6732b8487a4369960dcfaa6cc8f1f65164 | [
"MIT"
] | null | null | null | 92.151899 | 10,992 | 0.838049 | [
[
[
"# Reading Survey Data\n(Sanna Tyrvainen 2021)\n\nCode to read the soft CIFAR-10 survey results\n\nsurvey_answers = a pickle file with a list of arrays of survey results and original CIFAR-10 labels \n\ndata_batch_1 = a pickle file of CIFAR-10 1/5 training dataset with a dictionary of \n * b'batch_label', = 'training batch 1 of 5'\n * b'labels' = CIFAR-10 label \n * b'data' = CIFAR-10 images\n * b'filenames' = CIFAR-10 image names \n\n",
"_____no_output_____"
]
],
[
[
"\n%matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pickle\nimport torch\n\n\ndef unpickle(file):\n with open(file, 'rb') as fo:\n dict = pickle.load(fo, encoding='bytes')\n return dict\n \ndef imagshow(img):\n plt.imshow(np.transpose(img, (1, 2, 0)))\n plt.show()\n\n",
"_____no_output_____"
],
[
"labels = unpickle('survey_answers');\nimgdict = unpickle('data_batch_1');\n\nimgdata = imgdict[b'data'];\nlabeldata = imgdict[b'labels'];\n\nclass_names = ('plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck') \n \nimages = imgdata.reshape(len(imgdata),3, 32,32)\n\n\nprint('Example:')\n\nii = 7\n\nprint('survey answer: ', labels[ii])\nimagshow(images[ii])\nprint(labeldata[ii], class_names[labels[ii][1]])\n",
"Example:\nsurvey answer: ([0, 0, 0, 0, 2, 0, 0, 4, 0, 0], 7)\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
d00203b3851d7f8ba7d4dd294b0e49d96a8ba0b5 | 17,018 | ipynb | Jupyter Notebook | Neuroseeker_Analysis.ipynb | atabakd/start_brain | 7901b55fbe2ce1b10bb7e0d2f4f8e9987fed12dd | [
"MIT"
] | null | null | null | Neuroseeker_Analysis.ipynb | atabakd/start_brain | 7901b55fbe2ce1b10bb7e0d2f4f8e9987fed12dd | [
"MIT"
] | null | null | null | Neuroseeker_Analysis.ipynb | atabakd/start_brain | 7901b55fbe2ce1b10bb7e0d2f4f8e9987fed12dd | [
"MIT"
] | null | null | null | 32.726923 | 120 | 0.562992 | [
[
[
"Import the necessary imports",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function, division, absolute_import\n\nimport tensorflow as tf\nfrom tensorflow.contrib import keras\n\nimport numpy as np\nimport os\nfrom sklearn import preprocessing\nfrom sklearn.metrics import confusion_matrix\nimport itertools\nimport cPickle #python 2.x\n#import _pickle as cPickle #python 3.x\nimport h5py\nfrom matplotlib import pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Now read the data",
"_____no_output_____"
]
],
[
[
"with h5py.File(\"NS_LP_DS.h5\", \"r\") as hf:\n LFP_features_train = hf[\"LFP_features_train\"][...]\n targets_train = hf[\"targets_train\"][...]\n speeds_train = hf[\"speeds_train\"][...]\n LFP_features_eval = hf[\"LFP_features_eval\"][...]\n targets_eval = hf[\"targets_eval\"][...]\n speeds_eval = hf[\"speeds_eval\"][...]",
"_____no_output_____"
]
],
[
[
"And make sure it looks ok",
"_____no_output_____"
]
],
[
[
"rand_sample = np.random.randint(LFP_features_eval.shape[0])\nfor i in range(LFP_features_train.shape[-1]):\n plt.figure(figsize=(20,7))\n plt_data = LFP_features_eval[rand_sample,:,i]\n plt.plot(np.arange(-0.5, 0., 0.5/plt_data.shape[0]), plt_data)\n plt.xlable(\"time\")\n plt.title(str(i))",
"_____no_output_____"
]
],
[
[
"Now we write some helper functions to easily select regions.",
"_____no_output_____"
]
],
[
[
"block = np.array([[2,4,6,8],[1,3,5,7]])\nchannels = np.concatenate([(block + i*8) for i in range(180)][::-1])\nbrain_regions = {'Parietal Cortex': 8000, 'Hypocampus CA1': 6230, 'Hypocampus DG': 5760, 'Thalamus LPMR': 4450,\n 'Thalamus Posterior': 3500, 'Thalamus VPM': 1930, 'SubThalamic': 1050}\nbrain_regions = {k:v//22.5 for k,v in brain_regions.iteritems()}\nused_channels = np.arange(9,1440,20, dtype=np.int16)[:-6]\nfor i in (729,749,1209,1229):\n used_channels = np.delete(used_channels, np.where(used_channels==i)[0])\n\n# for k,v in brain_regions.iteritems():\n# print(\"{0}: {1}\".format(k,v))\n \nchannels_dict = {'Parietal Cortex': np.arange(1096,1440, dtype=np.int16), \n 'Hypocampus CA1': np.arange(1016,1096, dtype=np.int16), \n 'Hypocampus DG': np.arange(784,1016, dtype=np.int16), \n 'Thalamus LPMR': np.arange(616,784, dtype=np.int16),\n 'Thalamus Posterior': np.arange(340,616, dtype=np.int16), \n 'Thalamus VPM': np.arange(184,340, dtype=np.int16), \n 'SubThalamic': np.arange(184, dtype=np.int16)}\nused_channels_dict = {k:list() for k in channels_dict.iterkeys()}\n# print(\"hello\")\nfor ch in used_channels:\n for key in channels_dict.iterkeys():\n if ch in channels_dict[key]:\n used_channels_dict[key].append(ch)",
"_____no_output_____"
],
[
"LFP_features_train_current = LFP_features_train\nLFP_features_eval_current = LFP_features_eval\n# current_channels = np.sort(used_channels_dict['Hypocampus CA1']+used_channels_dict['Hypocampus DG']+\\\n# used_channels_dict['Thalamus Posterior'])\n# current_idxs = np.array([np.where(ch==used_channels)[0] for ch in current_channels]).squeeze()\n# LFP_features_train_current = LFP_features_train[...,current_idxs]\n# LFP_features_eval_current = LFP_features_eval[...,current_idxs]",
"_____no_output_____"
]
],
[
[
"Create a call back to save the best validation accuracy",
"_____no_output_____"
]
],
[
[
"model_chk_path = 'my_model.hdf5'\nmcp = keras.callbacks.ModelCheckpoint(model_chk_path, monitor=\"val_acc\",\n save_best_only=True)",
"_____no_output_____"
]
],
[
[
"Below I have defined a couple of different network architectures to play with.",
"_____no_output_____"
]
],
[
[
"# try:\n# model = None\n# except NameError:\n# pass\n# decay = 1e-3\n# conv1d = keras.layers.Convolution1D\n# maxPool = keras.layers.MaxPool1D\n# model = keras.models.Sequential()\n# model.add(conv1d(64, 5, padding='same', strides=2, activation='relu', \n# kernel_regularizer=keras.regularizers.l2(decay),\n# input_shape=LFP_features_train.shape[1:]))\n# model.add(maxPool())\n# model.add(conv1d(128, 3, padding='same', strides=2, activation='relu', \n# kernel_regularizer=keras.regularizers.l2(decay)))\n# model.add(maxPool())\n# model.add(conv1d(128, 3, padding='same', strides=2, activation='relu', \n# kernel_regularizer=keras.regularizers.l2(decay)))\n# model.add(maxPool())\n# model.add(conv1d(128, 3, padding='same', strides=2, activation='relu', \n# kernel_regularizer=keras.regularizers.l2(decay)))\n# model.add(maxPool())\n# model.add(keras.layers.Flatten())\n# model.add(keras.layers.Dropout(rate=0.5))\n# model.add(keras.layers.Dense(2, activation='softmax', kernel_regularizer=keras.regularizers.l2(decay)))",
"_____no_output_____"
],
[
"# try:\n# model = None\n# except NameError:\n# pass\n# decay = 1e-3\n# conv1d = keras.layers.Convolution1D\n# maxPool = keras.layers.MaxPool1D\n# BN = keras.layers.BatchNormalization\n# Act = keras.layers.Activation('relu')\n# model = keras.models.Sequential()\n# model.add(conv1d(64, 5, padding='same', strides=2, \n# kernel_regularizer=keras.regularizers.l1_l2(decay),\n# input_shape=LFP_features_train_current.shape[1:]))\n# model.add(BN())\n# model.add(Act)\n# model.add(maxPool())\n# model.add(conv1d(128, 3, padding='same', strides=2,\n# kernel_regularizer=keras.regularizers.l1_l2(decay)))\n# model.add(BN())\n# model.add(Act)\n# model.add(maxPool())\n# model.add(conv1d(128, 3, padding='same', strides=2,\n# kernel_regularizer=keras.regularizers.l1_l2(decay)))\n# model.add(BN())\n# model.add(Act)\n# model.add(maxPool())\n# model.add(conv1d(128, 3, padding='same', strides=2,\n# kernel_regularizer=keras.regularizers.l1_l2(decay)))\n# model.add(BN())\n# model.add(Act)\n# model.add(maxPool())\n# model.add(keras.layers.Flatten())\n# model.add(keras.layers.Dropout(rate=0.5))\n# model.add(keras.layers.Dense(2, activation='softmax', kernel_regularizer=keras.regularizers.l2(decay)))",
"_____no_output_____"
],
[
"# try:\n# model = None\n# except NameError:\n# pass\n# decay = 1e-3\n# conv1d = keras.layers.Convolution1D\n# maxPool = keras.layers.MaxPool1D\n# model = keras.models.Sequential()\n# model.add(conv1d(33, 5, padding='same', activation='relu', kernel_regularizer=keras.regularizers.l2(decay),\n# input_shape=LFP_features_train.shape[1:]))\n# model.add(maxPool())\n# model.add(conv1d(33, 3, padding='same', activation='relu', kernel_regularizer=keras.regularizers.l2(decay)))\n# model.add(maxPool())\n# model.add(conv1d(16, 3, padding='same', activation='relu', kernel_regularizer=keras.regularizers.l2(decay)))\n# model.add(maxPool())\n# model.add(conv1d(4, 3, padding='same', activation='relu', kernel_regularizer=keras.regularizers.l2(decay)))\n# model.add(maxPool())\n# model.add(keras.layers.Flatten())\n# model.add(keras.layers.Dropout(rate=0.5))\n# model.add(keras.layers.Dense(2, activation='softmax', kernel_regularizer=keras.regularizers.l2(decay)))",
"_____no_output_____"
],
[
"try:\n model = None\nexcept NameError:\n pass\ndecay = 1e-3\nregul = keras.regularizers.l1(decay)\nconv1d = keras.layers.Convolution1D\nmaxPool = keras.layers.MaxPool1D\nBN = keras.layers.BatchNormalization\nAct = keras.layers.Activation('relu')\nmodel = keras.models.Sequential()\nmodel.add(keras.layers.Convolution1D(64, 5, padding='same', strides=2, \n kernel_regularizer=keras.regularizers.l1_l2(decay),\n input_shape=LFP_features_train_current.shape[1:]))\nmodel.add(keras.layers.BatchNormalization())\nmodel.add(keras.layers.Activation('relu'))\nmodel.add(keras.layers.MaxPool1D())\nmodel.add(keras.layers.Convolution1D(128, 3, padding='same', strides=2,\n kernel_regularizer=keras.regularizers.l1_l2(decay)))\nmodel.add(keras.layers.BatchNormalization())\nmodel.add(keras.layers.Activation('relu'))\n# model.add(keras.layers.MaxPool1D())\n# model.add(keras.layers.Convolution1D(128, 3, padding='same', strides=2,\n# kernel_regularizer=keras.regularizers.l1_l2(decay)))\n# model.add(keras.layers.BatchNormalization())\n# model.add(keras.layers.Activation('relu'))\n\n# # model.add(keras.layers.GlobalMaxPooling1D())\n# model.add(keras.layers.MaxPool1D())\n# model.add(keras.layers.Convolution1D(128, 3, padding='same', strides=2,\n# kernel_regularizer=keras.regularizers.l1_l2(decay)))\n# model.add(keras.layers.BatchNormalization())\n# model.add(keras.layers.Activation('relu'))\n# model.add(maxPool())\n# model.add(keras.layers.Flatten())\nmodel.add(keras.layers.GlobalMaxPooling1D())\nmodel.add(keras.layers.Dropout(rate=0.5))\nmodel.add(keras.layers.Dense(2, activation='softmax', kernel_regularizer=keras.regularizers.l1_l2(decay)))",
"_____no_output_____"
],
[
"model.compile(optimizer='Adam',\nloss='categorical_crossentropy',\nmetrics=['accuracy'])",
"_____no_output_____"
],
[
"model.summary()",
"_____no_output_____"
],
[
"history = model.fit(LFP_features_train_current,\n targets_train,\n epochs=20,\n batch_size=1024,\n validation_data=(LFP_features_eval_current, targets_eval),\n callbacks=[mcp])",
"_____no_output_____"
]
],
[
[
"Helper function for the confusion matrix",
"_____no_output_____"
]
],
[
[
"def plot_confusion_matrix(cm, classes,\n normalize=False,\n title='Confusion matrix',\n cmap=plt.cm.Blues):\n \"\"\"\n This function prints and plots the confusion matrix.\n Normalization can be applied by setting `normalize=True`.\n \"\"\"\n if normalize:\n cm = cm.astype('float') / np.maximum(cm.sum(axis=1)[:, np.newaxis],1.0)\n print(\"Normalized confusion matrix\")\n else:\n print('Confusion matrix, without normalization')\n\n print(cm)\n \n cm = (cm*1000).astype(np.int16)\n cm = np.multiply(cm, 0.1)\n plt.imshow(cm, interpolation='nearest', cmap=cmap)\n plt.title(title)\n plt.colorbar()\n tick_marks = np.arange(len(classes))\n plt.xticks(tick_marks, classes, rotation=45)\n plt.yticks(tick_marks, classes)\n thresh = cm.max() / 2.\n for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):\n plt.text(j, i, \"{0}%\".format(cm[i, j]),\n horizontalalignment=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n\n plt.tight_layout()\n plt.ylabel('True label')\n plt.xlabel('Predicted label')\n return plt.gcf()",
"_____no_output_____"
],
[
"class_names = ['go', 'stop']\nmodel.load_weights('my_model.hdf5')\ny_pred_initial = model.predict(LFP_features_eval)\ntargets_eval_1d = np.argmax(targets_eval, axis=1)\ny_pred = np.argmax(y_pred_initial, axis=1)\ncnf_matrix = confusion_matrix(targets_eval_1d, y_pred)\nnp.set_printoptions(precision=2)\nplt.figure()\nfig = plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True,\n title='Normalized confusion matrix')",
"_____no_output_____"
],
[
"wrong_idxs = np.where(y_pred != targets_eval_1d)[0]\nwrong_vals = speeds_eval[wrong_idxs]\n# wrong_vals.squeeze().shape\n# crazy_wrong_idxs.shape",
"_____no_output_____"
],
[
"plt.cla()\nplt.close()\nplt.figure(figsize=(20,7))\nn, bins, patches = plt.hist(wrong_vals.squeeze(), \n bins=np.arange(0,1,0.01),)\n\nplt.plot(bins)\nplt.xlim([0,1])\nfig_dist = plt.gcf()",
"_____no_output_____"
]
],
[
[
"Train and evaluation accuracies",
"_____no_output_____"
]
],
[
[
"acc = history.history['acc']\nval_acc = history.history['val_acc']\nloss = history.history['loss']\nval_loss = history.history['val_loss']\nepochs = range(len(acc))\nplt.figure(figsize=(20,7))\nplt.plot(epochs, acc, 'bo', label='Training')\nplt.plot(epochs, val_acc, 'b', label='Validation')\nplt.title('Training and validation accuracy')\nplt.legend(loc='lower right', fontsize=24)\nplt.xticks(np.arange(20))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0020d0fb2d2788a3e363f47e1815235da9b2694 | 159,855 | ipynb | Jupyter Notebook | Instruction4/Instruction4-RegressionSVM.ipynb | danikhani/ITDS-Instructions-WS20 | f13691673a0ffc17f0ec2f4cdcaf588c90027116 | [
"MIT"
] | null | null | null | Instruction4/Instruction4-RegressionSVM.ipynb | danikhani/ITDS-Instructions-WS20 | f13691673a0ffc17f0ec2f4cdcaf588c90027116 | [
"MIT"
] | null | null | null | Instruction4/Instruction4-RegressionSVM.ipynb | danikhani/ITDS-Instructions-WS20 | f13691673a0ffc17f0ec2f4cdcaf588c90027116 | [
"MIT"
] | null | null | null | 165.139463 | 34,901 | 0.695562 | [
[
[
"# IDS Instruction: Regression\n(Lisa Mannel)",
"_____no_output_____"
],
[
"## Simple linear regression",
"_____no_output_____"
],
[
"First we import the packages necessary fo this instruction:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.metrics import mean_squared_error, mean_absolute_error",
"_____no_output_____"
]
],
[
[
"Consider the data set \"df\" with feature variables \"x\" and \"y\" given below.",
"_____no_output_____"
]
],
[
[
"df1 = pd.DataFrame({'x': [0, 1, 2, 3, 4, 5, 6, 7, 8, 9], 'y': [1, 3, 2, 5, 7, 8, 8, 9, 10, 12]})\nprint(df1)",
" x y\n0 0 1\n1 1 3\n2 2 2\n3 3 5\n4 4 7\n5 5 8\n6 6 8\n7 7 9\n8 8 10\n9 9 12\n"
]
],
[
[
"To get a first impression of the given data, let's have a look at its scatter plot:",
"_____no_output_____"
]
],
[
[
"plt.scatter(df1.x, df1.y, color = \"y\", marker = \"o\", s = 40)\nplt.xlabel('x') \nplt.ylabel('y')\nplt.title('first overview of the data')\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can already see a linear correlation between x and y. Assume the feature x to be descriptive, while y is our target feature. We want a linear function, y=ax+b, that predicts y as accurately as possible based on x. To achieve this goal we use linear regression from the sklearn package.",
"_____no_output_____"
]
],
[
[
"#define the set of descriptive features (in this case only 'x' is in that set) and the target feature (in this case 'y')\ndescriptiveFeatures1=df1[['x']]\nprint(descriptiveFeatures1)\ntargetFeature1=df1['y']\n\n#define the classifier\nclassifier = LinearRegression()\n#train the classifier\nmodel1 = classifier.fit(descriptiveFeatures1, targetFeature1)",
" x\n0 0\n1 1\n2 2\n3 3\n4 4\n5 5\n6 6\n7 7\n8 8\n9 9\n"
]
],
[
[
"Now we can use the classifier to predict y. We print the predictions as well as the coefficient and bias (*intercept*) of the linear function.",
"_____no_output_____"
]
],
[
[
"#use the classifier to make prediction\ntargetFeature1_predict = classifier.predict(descriptiveFeatures1)\nprint(targetFeature1_predict)\n#print coefficient and intercept\nprint('Coefficients: \\n', classifier.coef_)\nprint('Intercept: \\n', classifier.intercept_)",
"[ 1.23636364 2.40606061 3.57575758 4.74545455 5.91515152 7.08484848\n 8.25454545 9.42424242 10.59393939 11.76363636]\nCoefficients: \n [1.16969697]\nIntercept: \n 1.2363636363636399\n"
]
],
[
[
"Let's visualize our regression function with the scatterplot showing the original data set. Herefore, we use the predicted values.",
"_____no_output_____"
]
],
[
[
"#visualize data points\nplt.scatter(df1.x, df1.y, color = \"y\", marker = \"o\", s = 40) \n#visualize regression function\nplt.plot(descriptiveFeatures1, targetFeature1_predict, color = \"g\") \nplt.xlabel('x') \nplt.ylabel('y') \nplt.title('the data and the regression function')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### <span style=\"color:green\"> Now it is your turn. </span> Build a simple linear regression for the data below. Use col1 as descriptive feature and col2 as target feature. Also plot your results.",
"_____no_output_____"
]
],
[
[
"df2 = pd.DataFrame({'col1': [770, 677, 428, 410, 371, 504, 1136, 695, 551, 550], 'col2': [54, 47, 28, 38, 29, 38, 80, 52, 45, 40]})\n#Your turn\n# features that we use for the prediction are called the \"descriptive\" features\ndescriptiveFeatures2=df2[['col1']]\n# the feature we would like to predict is called target fueature\ntargetFeature2=df2['col2']\n\n# traing regression model: \nclassifier2 = LinearRegression()\nmodel2 = classifier2.fit(descriptiveFeatures2, targetFeature2)\n#use the classifier to make prediction\ntargetFeature2_predict = classifier2.predict(descriptiveFeatures2)\n\n#visualize data points\nplt.scatter(df2.col1, df2.col2, color = \"y\", marker = \"o\") \n#visualize regression function\nplt.plot(descriptiveFeatures2, targetFeature2_predict, color = \"g\") \nplt.xlabel('col1') \nplt.ylabel('col2') \nplt.title('the data and the regression function')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Evaluation\n\nUsually, the model and its predictions is not sufficient. In the following we want to evaluate our classifiers. \n\nLet's start by computing their error. The sklearn.metrics package contains several errors such as\n\n* Mean squared error\n* Mean absolute error\n* Mean squared log error\n* Median absolute error\n",
"_____no_output_____"
]
],
[
[
"#computing the squared error of the first model\nprint(\"Mean squared error model 1: %.2f\" % mean_squared_error(targetFeature1, targetFeature1_predict))",
"Mean squared error model 1: 0.56\n"
]
],
[
[
"We can also visualize the errors:",
"_____no_output_____"
]
],
[
[
"plt.scatter(targetFeature1_predict, (targetFeature1 - targetFeature1_predict) ** 2, color = \"blue\", s = 10,) \n \n## plotting line to visualize zero error \nplt.hlines(y = 0, xmin = 0, xmax = 15, linewidth = 2) \n \n## plot title \nplt.title(\"Squared errors Model 1\") \n \n## function to show plot \nplt.show() ",
"_____no_output_____"
]
],
[
[
"### <span style=\"color:green\"> Now it is your turn. </span> Compute the mean squared error and visualize the squared errors. Play around using different error metrics.",
"_____no_output_____"
]
],
[
[
"#Your turn\nprint(\"Mean squared error model 2: %.2f\" % mean_squared_error(targetFeature2,targetFeature2_predict))\nprint(\"Mean absolute error model 2: %.2f\" % mean_absolute_error(targetFeature2,targetFeature2_predict))\n\nplt.scatter(targetFeature2_predict, (targetFeature2 - targetFeature2_predict) ** 2, color = \"blue\",) \nplt.scatter(targetFeature2,abs(targetFeature2 - targetFeature2_predict),color = \"red\")\n \n## plotting line to visualize zero error \nplt.hlines(y = 0, xmin = 0, xmax = 80, linewidth = 2) \n \n## plot title \nplt.title(\"errors Model 2\") \n \n## function to show plot \nplt.show() \n\n",
"Mean squared error model 2: 8.89\nMean absolute error model 2: 2.32\n"
]
],
[
[
"## Handling multiple descriptive features at once - Multiple linear regression\nIn most cases, we will have more than one descriptive feature . As an example we use an example data set of the scikit package. The dataset describes housing prices in Boston based on several attributes. Note, in this format the data is already split into descriptive features and a target feature.",
"_____no_output_____"
]
],
[
[
"from sklearn import datasets ## imports datasets from scikit-learn\ndf3 = datasets.load_boston()\n\n#The sklearn package provides the data splitted into a set of descriptive features and a target feature.\n#We can easily transform this format into the pandas data frame as used above.\ndescriptiveFeatures3 = pd.DataFrame(df3.data, columns=df3.feature_names)\ntargetFeature3 = pd.DataFrame(df3.target, columns=['target'])\nprint('Descriptive features:')\nprint(descriptiveFeatures3.head())\nprint('Target feature:')\nprint(targetFeature3.head())",
"Descriptive features:\n CRIM ZN INDUS CHAS NOX RM AGE DIS RAD TAX \\\n0 0.00632 18.0 2.31 0.0 0.538 6.575 65.2 4.0900 1.0 296.0 \n1 0.02731 0.0 7.07 0.0 0.469 6.421 78.9 4.9671 2.0 242.0 \n2 0.02729 0.0 7.07 0.0 0.469 7.185 61.1 4.9671 2.0 242.0 \n3 0.03237 0.0 2.18 0.0 0.458 6.998 45.8 6.0622 3.0 222.0 \n4 0.06905 0.0 2.18 0.0 0.458 7.147 54.2 6.0622 3.0 222.0 \n\n PTRATIO B LSTAT \n0 15.3 396.90 4.98 \n1 17.8 396.90 9.14 \n2 17.8 392.83 4.03 \n3 18.7 394.63 2.94 \n4 18.7 396.90 5.33 \nTarget feature:\n target\n0 24.0\n1 21.6\n2 34.7\n3 33.4\n4 36.2\n"
]
],
[
[
"To predict the housing price we will use a Multiple Linear Regression model. In Python this is very straightforward: we use the same function as for simple linear regression, but our set of descriptive features now contains more than one element (see above).",
"_____no_output_____"
]
],
[
[
"classifier = LinearRegression()\nmodel3 = classifier.fit(descriptiveFeatures3,targetFeature3)\n\ntargetFeature3_predict = classifier.predict(descriptiveFeatures3)\nprint('Coefficients: \\n', classifier.coef_)\nprint('Intercept: \\n', classifier.intercept_)\nprint(\"Mean squared error: %.2f\" % mean_squared_error(targetFeature3, targetFeature3_predict))",
"Coefficients: \n [[-1.08011358e-01 4.64204584e-02 2.05586264e-02 2.68673382e+00\n -1.77666112e+01 3.80986521e+00 6.92224640e-04 -1.47556685e+00\n 3.06049479e-01 -1.23345939e-02 -9.52747232e-01 9.31168327e-03\n -5.24758378e-01]]\nIntercept: \n [36.45948839]\nMean squared error: 21.89\n"
]
],
[
[
"As you can see above, we have a coefficient for each descriptive feature.",
"_____no_output_____"
],
[
"## Handling categorical descriptive features\nSo far we always encountered numerical dscriptive features, but data sets can also contain categorical attributes. The regression function can only handle numerical input. There are several ways to tranform our categorical data to numerical data (for example using one-hot encoding as explained in the lecture: we introduce a 0/1 feature for every possible value of our categorical attribute). For adequate data, another possibility is to replace each categorical value by a numerical value and adding an ordering with it. \n\nPopular possibilities to achieve this transformation are\n\n* the get_dummies function of pandas\n* the OneHotEncoder of scikit\n* the LabelEncoder of scikit\n\nAfter encoding the attributes we can apply our regular regression function.",
"_____no_output_____"
]
],
[
[
"#example using pandas\ndf4 = pd.DataFrame({'A':['a','b','c'],'B':['c','b','a'] })\none_hot_pd = pd.get_dummies(df4)\none_hot_pd",
"_____no_output_____"
],
[
"#example using scikit\nfrom sklearn.preprocessing import LabelEncoder, OneHotEncoder\n\n#apply the one hot encoder\nencoder = OneHotEncoder(categories='auto')\nencoder.fit(df4)\ndf4_OneHot = encoder.transform(df4).toarray()\nprint('Transformed by One-hot Encoding: ')\nprint(df4_OneHot)\n\n# encode labels with value between 0 and n_classes-1\nencoder = LabelEncoder()\ndf4_LE = df4.apply(encoder.fit_transform)\nprint('Replacing categories by numerical labels: ')\nprint(df4_LE.head())",
"Transformed by One-hot Encoding: \n[[1. 0. 0. 0. 0. 1.]\n [0. 1. 0. 0. 1. 0.]\n [0. 0. 1. 1. 0. 0.]]\nReplacing categories by numerical labels: \n A B\n0 0 2\n1 1 1\n2 2 0\n"
]
],
[
[
"### <span style=\"color:green\"> Now it is your turn. </span> Perform linear regression using the data set given below. Don't forget to transform your categorical descriptive features. The rental price attribute represents the target variable. ",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import LabelEncoder\n\ndf5 = pd.DataFrame({'Size':[500,550,620,630,665],'Floor':[4,7,9,5,8], 'Energy rating':['C', 'A', 'A', 'B', 'C'], 'Rental price': [320,380,400,390,385] })\n#Your turn\n# To transform the categorial feature\nto_trannsform = df5[['Energy rating']]\nencoder = LabelEncoder()\ntransformed = to_trannsform.apply(encoder.fit_transform)\ndf5_transformed = df5\ndf5_transformed[['Energy rating']] = transformed\n\n# the feature we would like to predict is called target fueature\ndf5_traget = df5_transformed['Rental price']\n\n# features that we use for the prediction are called the \"descriptive\" features\ndf5_descpritive = df5_transformed[['Size','Floor','Energy rating']]\n\n# traing regression model: \nclassifier5 = LinearRegression()\nmodel5 = classifier5.fit(df5_descpritive, df5_traget)\n#use the classifier to make prediction\ntargetFeature5_predict = classifier5.predict(df5_descpritive)\n\nprint('Coefficients: \\n', classifier5.coef_)\nprint('Intercept: \\n', classifier5.intercept_)\nprint(\"Mean squared error: %.2f\" % mean_squared_error(df5_traget, targetFeature5_predict))\n\n",
"Coefficients: \n [ 0.39008474 -0.54300185 -18.80539593]\nIntercept: \n 166.068958800039\nMean squared error: 4.68\n"
]
],
[
[
"## Predicting a categorical target value - Logistic regression",
"_____no_output_____"
],
[
"We might also encounter data sets where our target feature is categorical. Here we don't transform them into numerical values, but insetad we use a logistic regression function. Luckily, sklearn provides us with a suitable function that is similar to the linear equivalent. Similar to linear regression, we can compute logistic regression on a single descriptive variable as well as on multiple variables.",
"_____no_output_____"
]
],
[
[
"# Importing the dataset\niris = pd.read_csv('iris.csv')\n\nprint('First look at the data set: ')\nprint(iris.head())\n\n#defining the descriptive and target features\ndescriptiveFeatures_iris = iris[['sepal_length']] #we only use the attribute 'sepal_length' in this example\ntargetFeature_iris = iris['species'] #we want to predict the 'species' of iris\n\nfrom sklearn.linear_model import LogisticRegression\nclassifier = LogisticRegression(solver = 'liblinear', multi_class = 'ovr')\nclassifier.fit(descriptiveFeatures_iris, targetFeature_iris)\n\ntargetFeature_iris_pred = classifier.predict(descriptiveFeatures_iris)\n\nprint('Coefficients: \\n', classifier.coef_)\nprint('Intercept: \\n', classifier.intercept_)",
"First look at the data set: \n sepal_length sepal_width petal_length petal_width species\n0 5.1 3.5 1.4 0.2 setosa\n1 4.9 3.0 1.4 0.2 setosa\n2 4.7 3.2 1.3 0.2 setosa\n3 4.6 3.1 1.5 0.2 setosa\n4 5.0 3.6 1.4 0.2 setosa\nCoefficients: \n [[-0.86959145]\n [ 0.01223362]\n [ 0.57972675]]\nIntercept: \n [ 4.16186636 -0.74244291 -3.9921824 ]\n"
]
],
[
[
"### <span style=\"color:green\"> Now it is your turn. </span> In the example above we only used the first attribute as descriptive variable. Change the example such that all available attributes are used.",
"_____no_output_____"
]
],
[
[
"#Your turn\n# Importing the dataset\niris2 = pd.read_csv('iris.csv')\n\nprint('First look at the data set: ')\nprint(iris2.head())\n\n#defining the descriptive and target features\ndescriptiveFeatures_iris2 = iris[['sepal_length','sepal_width','petal_length','petal_width']] \ntargetFeature_iris2 = iris['species'] #we want to predict the 'species' of iris\n\nfrom sklearn.linear_model import LogisticRegression\nclassifier2 = LogisticRegression(solver = 'liblinear', multi_class = 'ovr')\nclassifier2.fit(descriptiveFeatures_iris2, targetFeature_iris2)\n\ntargetFeature_iris_pred2 = classifier2.predict(descriptiveFeatures_iris2)\n\nprint('Coefficients: \\n', classifier2.coef_)\nprint('Intercept: \\n', classifier2.intercept_)",
"First look at the data set: \n sepal_length sepal_width petal_length petal_width species\n0 5.1 3.5 1.4 0.2 setosa\n1 4.9 3.0 1.4 0.2 setosa\n2 4.7 3.2 1.3 0.2 setosa\n3 4.6 3.1 1.5 0.2 setosa\n4 5.0 3.6 1.4 0.2 setosa\nCoefficients: \n [[ 0.41021713 1.46416217 -2.26003266 -1.02103509]\n [ 0.4275087 -1.61211605 0.5758173 -1.40617325]\n [-1.70751526 -1.53427768 2.47096755 2.55537041]]\nIntercept: \n [ 0.26421853 1.09392467 -1.21470917]\n"
]
],
[
[
"Note, that the regression classifier (both logistic and non-logistic) can be tweaked using several parameters. This includes, but is not limited to, non-linear regression. Check out the documentation for details and feel free to play around!",
"_____no_output_____"
],
[
"# Support Vector Machines",
"_____no_output_____"
],
[
"Aside from regression models, the sklearn package also provides us with a function for training support vector machines. Looking at the example below we see that they can be trained in similar ways. We still use the iris data set for illustration.",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import SVC\n\n#define descriptive and target features as before\ndescriptiveFeatures_iris = iris[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']]\ntargetFeature_iris = iris['species']\n\n#this time, we train an SVM classifier\nclassifier = SVC(C=1, kernel='linear', gamma = 'auto')\nclassifier.fit(descriptiveFeatures_iris, targetFeature_iris)\n\ntargetFeature_iris_predict = classifier.predict(descriptiveFeatures_iris)\ntargetFeature_iris_predict[0:5] #show the first 5 predicted values",
"_____no_output_____"
]
],
[
[
"As explained in the lecture, a support vector machine is defined by its support vectors. In the sklearn package we can access them and their properties very easily:\n\n* support_: indicies of support vectors\n* support_vectors_: the support vectors\n* n_support_: the number of support vectors for each class",
"_____no_output_____"
]
],
[
[
"print('Indicies of support vectors:')\nprint(classifier.support_)\n\nprint('The support vectors:')\nprint(classifier.support_vectors_)\n\nprint('The number of support vectors for each class:')\nprint(classifier.n_support_)",
"Indicies of support vectors:\n[ 23 24 41 52 56 63 66 68 70 72 76 77 83 84 98 106 110 119\n 123 126 127 129 133 138 146 147 149]\nThe support vectors:\n[[5.1 3.3 1.7 0.5]\n [4.8 3.4 1.9 0.2]\n [4.5 2.3 1.3 0.3]\n [6.9 3.1 4.9 1.5]\n [6.3 3.3 4.7 1.6]\n [6.1 2.9 4.7 1.4]\n [5.6 3. 4.5 1.5]\n [6.2 2.2 4.5 1.5]\n [5.9 3.2 4.8 1.8]\n [6.3 2.5 4.9 1.5]\n [6.8 2.8 4.8 1.4]\n [6.7 3. 5. 1.7]\n [6. 2.7 5.1 1.6]\n [5.4 3. 4.5 1.5]\n [5.1 2.5 3. 1.1]\n [4.9 2.5 4.5 1.7]\n [6.5 3.2 5.1 2. ]\n [6. 2.2 5. 1.5]\n [6.3 2.7 4.9 1.8]\n [6.2 2.8 4.8 1.8]\n [6.1 3. 4.9 1.8]\n [7.2 3. 5.8 1.6]\n [6.3 2.8 5.1 1.5]\n [6. 3. 4.8 1.8]\n [6.3 2.5 5. 1.9]\n [6.5 3. 5.2 2. ]\n [5.9 3. 5.1 1.8]]\nThe number of support vectors for each class:\n[ 3 12 12]\n"
]
],
[
[
"We can also calculate the distance of the data points to the separating hyperplane by using the decision_function(X) method. Score(X,y) calculates the mean accuracy of the classification. The classification report shows metrics such as precision, recall, f1-score and support. You will learn more about these quality metrics in a few lectures.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report\nclassifier.decision_function(descriptiveFeatures_iris)\nprint('Accuracy: \\n', classifier.score(descriptiveFeatures_iris,targetFeature_iris))\nprint('Classification report: \\n')\nprint(classification_report(targetFeature_iris, targetFeature_iris_predict)) ",
"Accuracy: \n 0.9933333333333333\nClassification report: \n\n precision recall f1-score support\n\n setosa 1.00 1.00 1.00 50\n versicolor 1.00 0.98 0.99 50\n virginica 0.98 1.00 0.99 50\n\n accuracy 0.99 150\n macro avg 0.99 0.99 0.99 150\nweighted avg 0.99 0.99 0.99 150\n\n"
]
],
[
[
"The SVC has many parameters. In the lecture you learned about the concept of kernels. Scikit gives you the opportunity to try different kernel functions.\nFurthermore, the parameter C tells the SVM optimization problem how much you want to avoid misclassifying each training example. ",
"_____no_output_____"
],
[
"On the scikit website you can find more information about the available kernels etc. http://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d002161c8340e231abaa9ca4c9f4b2c0350581f8 | 117,329 | ipynb | Jupyter Notebook | breast_linearsvm.ipynb | baopuzi/Breast_Cancer_Detection | 6b27ee4958c6eda4388830f316cca2fe343748ca | [
"MIT"
] | null | null | null | breast_linearsvm.ipynb | baopuzi/Breast_Cancer_Detection | 6b27ee4958c6eda4388830f316cca2fe343748ca | [
"MIT"
] | null | null | null | breast_linearsvm.ipynb | baopuzi/Breast_Cancer_Detection | 6b27ee4958c6eda4388830f316cca2fe343748ca | [
"MIT"
] | null | null | null | 392.404682 | 95,646 | 0.887129 | [
[
[
"# -*- coding: utf-8 -*-\n# 乳腺癌诊断分类\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sklearn.model_selection import train_test_split\nfrom sklearn import svm\nfrom sklearn import metrics\nfrom sklearn.preprocessing import StandardScaler\n\n# 加载数据集,你需要把数据放到目录中\ndata = pd.read_csv(\"./data/data.csv\")\n\n# 数据探索\n# 因为数据集中列比较多,我们需要把dataframe中的列全部显示出来\npd.set_option('display.max_columns', None)\nprint(data.columns)\nprint(data.head(5))\nprint(data.describe())\n\n# 将特征字段分成3组\nfeatures_mean= list(data.columns[2:12])\nfeatures_se= list(data.columns[12:22])\nfeatures_worst=list(data.columns[22:32])\n\n# 数据清洗\n# ID列没有用,删除该列\ndata.drop(\"id\",axis=1,inplace=True)\n# 将B良性替换为0,M恶性替换为1\ndata['diagnosis']=data['diagnosis'].map({'M':1,'B':0})\n\n# 将肿瘤诊断结果可视化\nsns.countplot(data['diagnosis'],label=\"Count\")\nplt.show()\n# 用热力图呈现features_mean字段之间的相关性\ncorr = data[features_mean].corr()\nplt.figure(figsize=(14,14))\n# annot=True显示每个方格的数据\nsns.heatmap(corr, annot=True)\nplt.show()\n\n\n# 特征选择\n#features_remain = ['radius_mean','texture_mean', 'smoothness_mean','compactness_mean','symmetry_mean', 'fractal_dimension_mean'] \nfeatures_remain = data.columns[1:31]\nprint(features_remain)\nprint('-'*100)\n# 抽取30%的数据作为测试集,其余作为训练集\ntrain, test = train_test_split(data, test_size = 0.3)# in this our main data is splitted into train and test\n# 抽取特征选择的数值作为训练和测试数据\ntrain_X = train[features_remain]\ntrain_y=train['diagnosis']\ntest_X= test[features_remain]\ntest_y =test['diagnosis']\n\n# 采用Z-Score规范化数据,保证每个特征维度的数据均值为0,方差为1\nss = StandardScaler()\ntrain_X = ss.fit_transform(train_X)\ntest_X = ss.transform(test_X)\n\n# 创建SVM分类器\nmodel = svm.LinearSVC()\n# 用训练集做训练\nmodel.fit(train_X,train_y)\n# 用测试集做预测\nprediction=model.predict(test_X)\nprint('准确率: ', metrics.accuracy_score(prediction,test_y))",
"Index(['id', 'diagnosis', 'radius_mean', 'texture_mean', 'perimeter_mean',\n 'area_mean', 'smoothness_mean', 'compactness_mean', 'concavity_mean',\n 'concave points_mean', 'symmetry_mean', 'fractal_dimension_mean',\n 'radius_se', 'texture_se', 'perimeter_se', 'area_se', 'smoothness_se',\n 'compactness_se', 'concavity_se', 'concave points_se', 'symmetry_se',\n 'fractal_dimension_se', 'radius_worst', 'texture_worst',\n 'perimeter_worst', 'area_worst', 'smoothness_worst',\n 'compactness_worst', 'concavity_worst', 'concave points_worst',\n 'symmetry_worst', 'fractal_dimension_worst'],\n dtype='object')\n id diagnosis radius_mean texture_mean perimeter_mean area_mean \\\n0 842302 M 17.99 10.38 122.80 1001.0 \n1 842517 M 20.57 17.77 132.90 1326.0 \n2 84300903 M 19.69 21.25 130.00 1203.0 \n3 84348301 M 11.42 20.38 77.58 386.1 \n4 84358402 M 20.29 14.34 135.10 1297.0 \n\n smoothness_mean compactness_mean concavity_mean concave points_mean \\\n0 0.11840 0.27760 0.3001 0.14710 \n1 0.08474 0.07864 0.0869 0.07017 \n2 0.10960 0.15990 0.1974 0.12790 \n3 0.14250 0.28390 0.2414 0.10520 \n4 0.10030 0.13280 0.1980 0.10430 \n\n symmetry_mean fractal_dimension_mean radius_se texture_se perimeter_se \\\n0 0.2419 0.07871 1.0950 0.9053 8.589 \n1 0.1812 0.05667 0.5435 0.7339 3.398 \n2 0.2069 0.05999 0.7456 0.7869 4.585 \n3 0.2597 0.09744 0.4956 1.1560 3.445 \n4 0.1809 0.05883 0.7572 0.7813 5.438 \n\n area_se smoothness_se compactness_se concavity_se concave points_se \\\n0 153.40 0.006399 0.04904 0.05373 0.01587 \n1 74.08 0.005225 0.01308 0.01860 0.01340 \n2 94.03 0.006150 0.04006 0.03832 0.02058 \n3 27.23 0.009110 0.07458 0.05661 0.01867 \n4 94.44 0.011490 0.02461 0.05688 0.01885 \n\n symmetry_se fractal_dimension_se radius_worst texture_worst \\\n0 0.03003 0.006193 25.38 17.33 \n1 0.01389 0.003532 24.99 23.41 \n2 0.02250 0.004571 23.57 25.53 \n3 0.05963 0.009208 14.91 26.50 \n4 0.01756 0.005115 22.54 16.67 \n\n perimeter_worst area_worst smoothness_worst compactness_worst \\\n0 184.60 2019.0 0.1622 0.6656 \n1 158.80 1956.0 0.1238 0.1866 \n2 152.50 1709.0 0.1444 0.4245 \n3 98.87 567.7 0.2098 0.8663 \n4 152.20 1575.0 0.1374 0.2050 \n\n concavity_worst concave points_worst symmetry_worst \\\n0 0.7119 0.2654 0.4601 \n1 0.2416 0.1860 0.2750 \n2 0.4504 0.2430 0.3613 \n3 0.6869 0.2575 0.6638 \n4 0.4000 0.1625 0.2364 \n\n fractal_dimension_worst \n0 0.11890 \n1 0.08902 \n2 0.08758 \n3 0.17300 \n4 0.07678 \n id radius_mean texture_mean perimeter_mean area_mean \\\ncount 5.690000e+02 569.000000 569.000000 569.000000 569.000000 \nmean 3.037183e+07 14.127292 19.289649 91.969033 654.889104 \nstd 1.250206e+08 3.524049 4.301036 24.298981 351.914129 \nmin 8.670000e+03 6.981000 9.710000 43.790000 143.500000 \n25% 8.692180e+05 11.700000 16.170000 75.170000 420.300000 \n50% 9.060240e+05 13.370000 18.840000 86.240000 551.100000 \n75% 8.813129e+06 15.780000 21.800000 104.100000 782.700000 \nmax 9.113205e+08 28.110000 39.280000 188.500000 2501.000000 \n\n smoothness_mean compactness_mean concavity_mean concave points_mean \\\ncount 569.000000 569.000000 569.000000 569.000000 \nmean 0.096360 0.104341 0.088799 0.048919 \nstd 0.014064 0.052813 0.079720 0.038803 \nmin 0.052630 0.019380 0.000000 0.000000 \n25% 0.086370 0.064920 0.029560 0.020310 \n50% 0.095870 0.092630 0.061540 0.033500 \n75% 0.105300 0.130400 0.130700 0.074000 \nmax 0.163400 0.345400 0.426800 0.201200 \n\n symmetry_mean fractal_dimension_mean radius_se texture_se \\\ncount 569.000000 569.000000 569.000000 569.000000 \nmean 0.181162 0.062798 0.405172 1.216853 \nstd 0.027414 0.007060 0.277313 0.551648 \nmin 0.106000 0.049960 0.111500 0.360200 \n25% 0.161900 0.057700 0.232400 0.833900 \n50% 0.179200 0.061540 0.324200 1.108000 \n75% 0.195700 0.066120 0.478900 1.474000 \nmax 0.304000 0.097440 2.873000 4.885000 \n\n perimeter_se area_se smoothness_se compactness_se concavity_se \\\ncount 569.000000 569.000000 569.000000 569.000000 569.000000 \nmean 2.866059 40.337079 0.007041 0.025478 0.031894 \nstd 2.021855 45.491006 0.003003 0.017908 0.030186 \nmin 0.757000 6.802000 0.001713 0.002252 0.000000 \n25% 1.606000 17.850000 0.005169 0.013080 0.015090 \n50% 2.287000 24.530000 0.006380 0.020450 0.025890 \n75% 3.357000 45.190000 0.008146 0.032450 0.042050 \nmax 21.980000 542.200000 0.031130 0.135400 0.396000 \n\n concave points_se symmetry_se fractal_dimension_se radius_worst \\\ncount 569.000000 569.000000 569.000000 569.000000 \nmean 0.011796 0.020542 0.003795 16.269190 \nstd 0.006170 0.008266 0.002646 4.833242 \nmin 0.000000 0.007882 0.000895 7.930000 \n25% 0.007638 0.015160 0.002248 13.010000 \n50% 0.010930 0.018730 0.003187 14.970000 \n75% 0.014710 0.023480 0.004558 18.790000 \nmax 0.052790 0.078950 0.029840 36.040000 \n\n texture_worst perimeter_worst area_worst smoothness_worst \\\ncount 569.000000 569.000000 569.000000 569.000000 \nmean 25.677223 107.261213 880.583128 0.132369 \nstd 6.146258 33.602542 569.356993 0.022832 \nmin 12.020000 50.410000 185.200000 0.071170 \n25% 21.080000 84.110000 515.300000 0.116600 \n50% 25.410000 97.660000 686.500000 0.131300 \n75% 29.720000 125.400000 1084.000000 0.146000 \nmax 49.540000 251.200000 4254.000000 0.222600 \n\n compactness_worst concavity_worst concave points_worst \\\ncount 569.000000 569.000000 569.000000 \nmean 0.254265 0.272188 0.114606 \nstd 0.157336 0.208624 0.065732 \nmin 0.027290 0.000000 0.000000 \n25% 0.147200 0.114500 0.064930 \n50% 0.211900 0.226700 0.099930 \n75% 0.339100 0.382900 0.161400 \nmax 1.058000 1.252000 0.291000 \n\n symmetry_worst fractal_dimension_worst \ncount 569.000000 569.000000 \nmean 0.290076 0.083946 \nstd 0.061867 0.018061 \nmin 0.156500 0.055040 \n25% 0.250400 0.071460 \n50% 0.282200 0.080040 \n75% 0.317900 0.092080 \nmax 0.663800 0.207500 \n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d002285d31868aabc4f0896a5ec20210a71ba83c | 44,048 | ipynb | Jupyter Notebook | NASA/Python_codes/ML_Books/01_01_transfer_learning_model_EVI.ipynb | HNoorazar/Kirti | fb7108dac1190774bd90a527aaa8a3cb405f127d | [
"MIT"
] | null | null | null | NASA/Python_codes/ML_Books/01_01_transfer_learning_model_EVI.ipynb | HNoorazar/Kirti | fb7108dac1190774bd90a527aaa8a3cb405f127d | [
"MIT"
] | null | null | null | NASA/Python_codes/ML_Books/01_01_transfer_learning_model_EVI.ipynb | HNoorazar/Kirti | fb7108dac1190774bd90a527aaa8a3cb405f127d | [
"MIT"
] | null | null | null | 154.554386 | 35,908 | 0.894956 | [
[
[
"import numpy as np\nimport pandas as pd\nfrom datetime import date\nfrom random import seed\nfrom random import random\n\nimport time\nimport scipy, scipy.signal\nimport os, os.path\nimport shutil\nimport matplotlib\nimport matplotlib.pyplot as plt\n\nfrom pylab import imshow\n\n# vgg16 model used for transfer learning on the dogs and cats dataset\nfrom matplotlib import pyplot\n# from keras.utils import to_categorical\nfrom tensorflow.keras.utils import to_categorical\nfrom keras.models import Sequential\nfrom keras.applications.vgg16 import VGG16\nfrom keras.models import Model\nfrom keras.layers import Dense\nfrom keras.layers import Flatten\nimport tensorflow as tf\n# from keras.optimizers import SGD\n\nfrom keras.layers import Conv2D\nfrom keras.layers import MaxPooling2D\n\n# from keras.optimizers import gradient_descent_v2\n# SGD = gradient_descent_v2.SGD(...)\n\nfrom tensorflow.keras.optimizers import SGD\nfrom keras.preprocessing.image import ImageDataGenerator\n\n\nimport h5py\nimport sys\nsys.path.append('/Users/hn/Documents/00_GitHub/Ag/NASA/Python_codes/')\nimport NASA_core as nc\n# import NASA_plot_core.py as rcp",
"_____no_output_____"
],
[
"from keras.preprocessing.image import load_img\nfrom keras.preprocessing.image import img_to_array\nfrom keras.models import load_model",
"_____no_output_____"
],
[
"idx = \"EVI\"\ntrain_folder = '/Users/hn/Documents/01_research_data/NASA/ML_data/train_images_' + idx + '/'\ntest_folder = \"/Users/hn/Documents/01_research_data/NASA/ML_data/limitCrops_nonExpert_images/\"",
"_____no_output_____"
]
],
[
[
"# Prepare final dataset",
"_____no_output_____"
]
],
[
[
"# organize dataset into a useful structure\n# create directories\ndataset_home = train_folder\n\n# create label subdirectories\nlabeldirs = ['separate_singleDouble/single/', 'separate_singleDouble/double/']\nfor labldir in labeldirs:\n newdir = dataset_home + labldir\n os.makedirs(newdir, exist_ok=True)\n \n# copy training dataset images into subdirectories\nfor file in os.listdir(train_folder):\n src = train_folder + '/' + file\n if file.startswith('single'):\n dst = dataset_home + 'separate_singleDouble/single/' + file\n shutil.copyfile(src, dst)\n elif file.startswith('double'):\n dst = dataset_home + 'separate_singleDouble/double/' + file\n shutil.copyfile(src, dst)",
"_____no_output_____"
]
],
[
[
"# Plot For Fun",
"_____no_output_____"
]
],
[
[
"# plot dog photos from the dogs vs cats dataset\nfrom matplotlib.image import imread\n\n\n# define location of dataset\n# plot first few images\nfiles = os.listdir(train_folder)[2:4]\n# files = [sorted(os.listdir(train_folder))[2]] + [sorted(os.listdir(train_folder))[-2]]\nfor i in range(2):\n # define subplot\n pyplot.subplot(210 + 1 + i)\n # define filename\n filename = train_folder + files[i]\n # load image pixels\n image = imread(filename)\n # plot raw pixel data\n pyplot.imshow(image)\n# show the figure\npyplot.show()",
"_____no_output_____"
]
],
[
[
"# Full Code",
"_____no_output_____"
]
],
[
[
"# define cnn model\ndef define_model():\n # load model\n model = VGG16(include_top=False, input_shape=(224, 224, 3))\n # mark loaded layers as not trainable\n for layer in model.layers:\n layer.trainable = False\n # add new classifier layers\n flat1 = Flatten()(model.layers[-1].output)\n class1 = Dense(128, activation='relu', kernel_initializer='he_uniform')(flat1)\n output = Dense(1, activation='sigmoid')(class1)\n # define new model\n model = Model(inputs=model.inputs, outputs=output)\n # compile model\n opt = SGD(learning_rate=0.001, momentum=0.9)\n model.compile(optimizer=opt, loss='binary_crossentropy', metrics=['accuracy'])\n return model\n\n# run the test harness for evaluating a model\ndef run_test_harness():\n # define model\n _model = define_model()\n # create data generator\n datagen = ImageDataGenerator(featurewise_center=True)\n # specify imagenet mean values for centering\n datagen.mean = [123.68, 116.779, 103.939]\n # prepare iterator\n train_separate_dir = train_folder + \"separate_singleDouble/\"\n train_it = datagen.flow_from_directory(train_separate_dir,\n class_mode='binary', \n batch_size=16, \n target_size=(224, 224))\n # fit model\n _model.fit(train_it, \n steps_per_epoch=len(train_it), \n epochs=10, verbose=1)\n \n model_dir = \"/Users/hn/Documents/01_research_data/NASA/ML_Models/\"\n _model.save(model_dir+'01_TL_SingleDouble.h5')\n# tf.keras.models.save_model(model=trained_model, filepath=model_dir+'01_TL_SingleDouble.h5')\n \n# return(_model)\n\n# entry point, run the test harness\nstart_time = time.time()\nrun_test_harness()\nend_time = time.time()",
"_____no_output_____"
],
[
"# photo = load_img(train_folder + files[0], target_size=(200, 500))\n# photo",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0022fad4be435c8f50aaaaff8f8656636ca9192 | 48,824 | ipynb | Jupyter Notebook | Random Forest/RF_classification.ipynb | InternityFoundation/Machine-Learning-Akshat | 728ec2a4fe036bbd117b714f34f4cde0a61642bf | [
"Apache-2.0"
] | null | null | null | Random Forest/RF_classification.ipynb | InternityFoundation/Machine-Learning-Akshat | 728ec2a4fe036bbd117b714f34f4cde0a61642bf | [
"Apache-2.0"
] | null | null | null | Random Forest/RF_classification.ipynb | InternityFoundation/Machine-Learning-Akshat | 728ec2a4fe036bbd117b714f34f4cde0a61642bf | [
"Apache-2.0"
] | null | null | null | 106.13913 | 20,124 | 0.828691 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.cross_validation import train_test_split \nfrom sklearn.metrics import accuracy_score\nfrom sklearn.preprocessing import StandardScaler",
"_____no_output_____"
],
[
"data = pd.read_csv('Social_Network_Ads.csv')",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"data.isnull().sum()",
"_____no_output_____"
],
[
"from sklearn import preprocessing\nle = preprocessing.LabelEncoder()\nle.fit(['Male','Female'])\ndata['Gender']=le.transform(data['Gender'])\ndata.head()",
"_____no_output_____"
],
[
"X = data.iloc[:, 2:4].values\ny = data.iloc[:, 4].values\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.3)\nsc = StandardScaler()\nX_train = sc.fit_transform(X_train)\nX_test = sc.transform(X_test)\n",
"C:\\Users\\admin\\Anaconda3\\lib\\site-packages\\sklearn\\utils\\validation.py:475: DataConversionWarning: Data with input dtype int64 was converted to float64 by StandardScaler.\n warnings.warn(msg, DataConversionWarning)\n"
],
[
"from sklearn.ensemble import RandomForestClassifier\nclf = RandomForestClassifier(n_estimators = 100, criterion = 'gini')\nclf.fit(X_train, y_train)",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix\nconfusion_matrix(y_test, clf.predict(X_test))",
"_____no_output_____"
],
[
"confusion_matrix(y_train, clf.predict(X_train))",
"_____no_output_____"
],
[
"print (\"Testing Accuracy is : \",accuracy_score(y_test,clf.predict(X_test)))\nprint (\"Training Accuracy is : \",accuracy_score(y_train,clf.predict(X_train)))",
"Testing Accuracy is : 0.933333333333\nTraining Accuracy is : 0.992857142857\n"
],
[
"from matplotlib.colors import ListedColormap\nX_set, y_set = X_train, y_train\nX1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 1, stop = X_set[:, 0].max() + 1, step = 0.01),\n np.arange(start = X_set[:, 1].min() - 1, stop = X_set[:, 1].max() + 1, step = 0.01))\nplt.contourf(X1, X2, clf.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),\n alpha = 0.75, cmap = ListedColormap(('red', 'green')))\nplt.xlim(X1.min(), X1.max())\nplt.ylim(X2.min(), X2.max())\nfor i, j in enumerate(np.unique(y_set)):\n plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1],\n c = ListedColormap(('red', 'green'))(i), label = j)\nplt.title('Classifier (Training set)')\nplt.xlabel('Age')\nplt.ylabel('Estimated Salary')\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"from matplotlib.colors import ListedColormap\nX_set, y_set = X_test, y_test\nX1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 1, stop = X_set[:, 0].max() + 1, step = 0.01),\n np.arange(start = X_set[:, 1].min() - 1, stop = X_set[:, 1].max() + 1, step = 0.01))\nplt.contourf(X1, X2, clf.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),\n alpha = 0.75, cmap = ListedColormap(('red', 'green')))\nplt.xlim(X1.min(), X1.max())\nplt.ylim(X2.min(), X2.max())\nfor i, j in enumerate(np.unique(y_set)):\n plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1],\n c = ListedColormap(('red', 'green'))(i), label = j)\nplt.title('Classifier (Test set)')\nplt.xlabel('Age')\nplt.ylabel('Estimated Salary')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d002337a342addb363e0327a0e1d6d4c1e3a0c98 | 15,072 | ipynb | Jupyter Notebook | docs/Supplementary-Materials/01-Spark-SQL.ipynb | ymei9/Big-Data-Analytics-for-Business | fba226e86a47ff188562655ce23b7af79781948a | [
"MIT"
] | 12 | 2019-02-01T01:02:02.000Z | 2022-03-22T22:45:39.000Z | docs/Supplementary-Materials/01-Spark-SQL.ipynb | ymei9/Big-Data-Analytics-for-Business | fba226e86a47ff188562655ce23b7af79781948a | [
"MIT"
] | null | null | null | docs/Supplementary-Materials/01-Spark-SQL.ipynb | ymei9/Big-Data-Analytics-for-Business | fba226e86a47ff188562655ce23b7af79781948a | [
"MIT"
] | 16 | 2019-02-03T15:56:51.000Z | 2022-03-29T03:34:21.000Z | 27.756906 | 535 | 0.524151 | [
[
[
"# Spark SQL\n\nSpark SQL is arguably one of the most important and powerful features in Spark. In a nutshell, with Spark SQL you can run SQL queries against views or tables organized into databases. You also can use system functions or define user functions and analyze query plans in order to optimize their workloads. This integrates directly into the DataFrame API, and as we saw in previous classes, you can choose to express some of your data manipulations in SQL and others in DataFrames and they will compile to the same underlying code.",
"_____no_output_____"
],
[
"## Big Data and SQL: Apache Hive\n\nBefore Spark’s rise, Hive was the de facto big data SQL access layer. Originally developed at Facebook, Hive became an incredibly popular tool across industry for performing SQL operations on big data. In many ways it helped propel Hadoop into different industries because analysts could run SQL queries. Although Spark began as a general processing engine with Resilient Distributed Datasets (RDDs), a large cohort of users now use Spark SQL.\n\n## Big Data and SQL: Spark SQL\n\nWith the release of Spark 2.0, its authors created a superset of Hive’s support, writing a native SQL parser that supports both ANSI-SQL as well as HiveQL queries. This, along with its unique interoperability with DataFrames, makes it a powerful tool for all sorts of companies. For example, in late 2016, Facebook announced that it had begun running Spark workloads and seeing large benefits in doing so. In the words of the blog post’s authors:\n\n>We challenged Spark to replace a pipeline that decomposed to hundreds of Hive jobs into a single Spark job. Through a series of performance and reliability improvements, we were able to scale Spark to handle one of our entity ranking data processing use cases in production…. The Spark-based pipeline produced significant performance improvements (4.5–6x CPU, 3–4x resource reservation, and ~5x latency) compared with the old Hive-based pipeline, and it has been running in production for several months.\n\nThe power of Spark SQL derives from several key facts: SQL analysts can now take advantage of Spark’s computation abilities by plugging into the Thrift Server or Spark’s SQL interface, whereas data engineers and scientists can use Spark SQL where appropriate in any data flow. This unifying API allows for data to be extracted with SQL, manipulated as a DataFrame, passed into one of Spark MLlibs’ large-scale machine learning algorithms, written out to another data source, and everything in between.\n\n**NOTE:** Spark SQL is intended to operate as an online analytic processing (OLAP) database, not an online transaction processing (OLTP) database. This means that it is not intended to perform extremely low-latency queries. Even though support for in-place modifications is sure to be something that comes up in the future, it’s not something that is currently available.",
"_____no_output_____"
]
],
[
[
"spark.sql(\"SELECT 1 + 1\").show()",
"+-------+\n|(1 + 1)|\n+-------+\n| 2|\n+-------+\n\n"
]
],
[
[
"As we have seen before, you can completely interoperate between SQL and DataFrames, as you see fit. For instance, you can create a DataFrame, manipulate it with SQL, and then manipulate it again as a DataFrame. It’s a powerful abstraction that you will likely find yourself using quite a bit:",
"_____no_output_____"
]
],
[
[
"bucket = spark._jsc.hadoopConfiguration().get(\"fs.gs.system.bucket\")\ndata = \"gs://\" + bucket + \"/notebooks/data/\"\n\nspark.read.json(data + \"flight-data/json/2015-summary.json\")\\\n .createOrReplaceTempView(\"flights_view\") # DF => SQL",
"_____no_output_____"
],
[
"spark.sql(\"\"\"\nSELECT DEST_COUNTRY_NAME, sum(count)\nFROM flights_view GROUP BY DEST_COUNTRY_NAME\n\"\"\")\\\n .where(\"DEST_COUNTRY_NAME like 'S%'\").where(\"`sum(count)` > 10\")\\\n .count() # SQL => DF",
"_____no_output_____"
]
],
[
[
"## Creating Tables\n\nYou can create tables from a variety of sources. For instance below we are creating a table from a SELECT statement:",
"_____no_output_____"
]
],
[
[
"spark.sql('''\nCREATE TABLE IF NOT EXISTS flights_from_select USING parquet AS SELECT * FROM flights_view\n''')",
"_____no_output_____"
],
[
"spark.sql('SELECT * FROM flights_from_select').show(5)",
"+-----------------+-------------------+-----+\n|DEST_COUNTRY_NAME|ORIGIN_COUNTRY_NAME|count|\n+-----------------+-------------------+-----+\n| United States| Romania| 15|\n| United States| Croatia| 1|\n| United States| Ireland| 344|\n| Egypt| United States| 15|\n| United States| India| 62|\n+-----------------+-------------------+-----+\nonly showing top 5 rows\n\n"
],
[
"spark.sql('''\nDESCRIBE TABLE flights_from_select\n''').show()",
"+-------------------+---------+-------+\n| col_name|data_type|comment|\n+-------------------+---------+-------+\n| DEST_COUNTRY_NAME| string| null|\n|ORIGIN_COUNTRY_NAME| string| null|\n| count| bigint| null|\n+-------------------+---------+-------+\n\n"
]
],
[
[
"## Catalog\nThe highest level abstraction in Spark SQL is the Catalog. The Catalog is an abstraction for the storage of metadata about the data stored in your tables as well as other helpful things like databases, tables, functions, and views. The catalog is available in the `spark.catalog` package and contains a number of helpful functions for doing things like listing tables, databases, and functions.",
"_____no_output_____"
]
],
[
[
"Cat = spark.catalog",
"_____no_output_____"
],
[
"Cat.listTables()",
"_____no_output_____"
],
[
"spark.sql('SHOW TABLES').show(5, False)",
"+--------+-------------------+-----------+\n|database|tableName |isTemporary|\n+--------+-------------------+-----------+\n|default |flights_from_select|false |\n| |flights_view |true |\n+--------+-------------------+-----------+\n\n"
],
[
"Cat.listDatabases()",
"_____no_output_____"
],
[
"spark.sql('SHOW DATABASES').show()",
"+------------+\n|databaseName|\n+------------+\n| default|\n+------------+\n\n"
],
[
"Cat.listColumns('flights_from_select')",
"_____no_output_____"
],
[
"Cat.listTables()",
"_____no_output_____"
]
],
[
[
"### Caching Tables",
"_____no_output_____"
]
],
[
[
"spark.sql('''\nCACHE TABLE flights_view\n''')",
"_____no_output_____"
],
[
"spark.sql('''\nUNCACHE TABLE flights_view\n''')",
"_____no_output_____"
]
],
[
[
"## Explain",
"_____no_output_____"
]
],
[
[
"spark.sql('''\nEXPLAIN SELECT * FROM just_usa_view\n''').show(1, False)",
"+-----------------------------------------------------------------------------------------------------------------+\n|plan |\n+-----------------------------------------------------------------------------------------------------------------+\n|== Physical Plan ==\norg.apache.spark.sql.AnalysisException: Table or view not found: just_usa_view; line 2 pos 22|\n+-----------------------------------------------------------------------------------------------------------------+\n\n"
]
],
[
[
"### VIEWS - create/drop",
"_____no_output_____"
]
],
[
[
"spark.sql('''\nCREATE VIEW just_usa_view AS\n SELECT * FROM flights_from_select WHERE dest_country_name = 'United States'\n''')",
"_____no_output_____"
],
[
"spark.sql('''\nDROP VIEW IF EXISTS just_usa_view\n''')",
"_____no_output_____"
]
],
[
[
"### Drop tables",
"_____no_output_____"
]
],
[
[
"spark.sql('DROP TABLE flights_from_select')",
"_____no_output_____"
],
[
"spark.sql('DROP TABLE IF EXISTS flights_from_select')",
"_____no_output_____"
]
],
[
[
"## `spark-sql`\n\nGo to the command line tool and check for the list of databases and tables. For instance:\n\n`SHOW TABLES`",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d002549fdfb1e35d86637b3c9d97a47a8f614a9f | 17,252 | ipynb | Jupyter Notebook | .ipynb_checkpoints/LaneDetect-checkpoint.ipynb | Eng-Mo/CarND-Advanced-Lane-Lines | 1fc98e892f22ecdae81e1b02b10335be5eabcd88 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/LaneDetect-checkpoint.ipynb | Eng-Mo/CarND-Advanced-Lane-Lines | 1fc98e892f22ecdae81e1b02b10335be5eabcd88 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/LaneDetect-checkpoint.ipynb | Eng-Mo/CarND-Advanced-Lane-Lines | 1fc98e892f22ecdae81e1b02b10335be5eabcd88 | [
"MIT"
] | 1 | 2020-04-21T10:50:43.000Z | 2020-04-21T10:50:43.000Z | 38.508929 | 161 | 0.546719 | [
[
[
"import numpy as np\nimport cv2\nimport matplotlib\nimport matplotlib.pyplot as plt\nimport matplotlib as mpimg\nimport numpy as np\nfrom IPython.display import HTML\nimport os, sys\nimport glob\nimport moviepy\nfrom moviepy.editor import VideoFileClip\nfrom moviepy.editor import * \nfrom IPython import display\nfrom IPython.core.display import display\nfrom IPython.display import Image\nimport pylab\nimport scipy.misc\n\n",
"_____no_output_____"
],
[
"def region_of_interest(img):\n mask = np.zeros(img.shape, dtype=np.uint8) #mask image\n roi_corners = np.array([[(200,675), (1200,675), (700,430),(500,430)]], \n dtype=np.int32) # vertisies seted to form trapezoidal scene\n channel_count = 1#img.shape[2] # image channels\n ignore_mask_color = (255,)*channel_count\n cv2.fillPoly(mask, roi_corners, ignore_mask_color) \n masked_image = cv2.bitwise_and(img, mask)\n \n return masked_image\n\n",
"_____no_output_____"
],
[
"def ColorThreshold(img): # Threshold Yellow anf White Colos from RGB, HSV, HLS color spaces\n \n HSV = cv2.cvtColor(img, cv2.COLOR_RGB2HSV)\n\n # For yellow\n yellow = cv2.inRange(HSV, (20, 100, 100), (50, 255, 255))\n\n # For white\n sensitivity_1 = 68\n white = cv2.inRange(HSV, (0,0,255-sensitivity_1), (255,20,255))\n\n sensitivity_2 = 60\n HSL = cv2.cvtColor(img, cv2.COLOR_RGB2HLS)\n white_2 = cv2.inRange(HSL, (0,255-sensitivity_2,0), (255,255,sensitivity_2))\n white_3 = cv2.inRange(img, (200,200,200), (255,255,255))\n\n bit_layer = yellow | white | white_2 | white_3\n \n return bit_layer\n\n",
"_____no_output_____"
],
[
"from skimage import morphology\n\ndef SobelThr(img): # Sobel edge detection extraction\n gray=img\n \n \n sobelx = cv2.Sobel(gray, cv2.CV_64F, 1, 0,ksize=15)\n sobely = cv2.Sobel(gray, cv2.CV_64F, 0, 1,ksize=15)\n \n abs_sobelx = np.absolute(sobelx)\n abs_sobely = np.absolute(sobely)\n scaled_sobelx = np.uint8(255*abs_sobelx/np.max(abs_sobelx))\n scaled_sobely = np.uint8(255*abs_sobely/np.max(abs_sobely))\n \n \n binary_outputabsx = np.zeros_like(scaled_sobelx)\n binary_outputabsx[(scaled_sobelx >= 70) & (scaled_sobelx <= 255)] = 1\n \n \n \n binary_outputabsy = np.zeros_like(scaled_sobely)\n binary_outputabsy[(scaled_sobely >= 100) & (scaled_sobely <= 150)] = 1\n\n \n mag_thresh=(100, 200)\n gradmag = np.sqrt(sobelx**2 + sobely**2)\n \n\n scale_factor = np.max(gradmag)/255\n gradmag = (gradmag/scale_factor).astype(np.uint8) \n binary_outputmag = np.zeros_like(gradmag)\n binary_outputmag[(gradmag >= mag_thresh[0]) & (gradmag <= mag_thresh[1])] = 1\n combinedS = np.zeros_like(binary_outputabsx)\n combinedS[(((binary_outputabsx == 1) | (binary_outputabsy == 1))|(binary_outputmag==1)) ] = 1\n \n return combinedS\n\n",
"_____no_output_____"
],
[
"def combinI(b1,b2): ##Combine color threshold + Sobel edge detection\n\n combined = np.zeros_like(b1)\n combined[((b1 == 1)|(b2 == 255)) ] = 1\n\n \n return combined",
"_____no_output_____"
],
[
"def prespectI(img): # Calculate the prespective transform and warp the Image to the eye bird view\n \n \n\n src=np.float32([[728,475],\n [1058,690],\n [242,690],\n [565,475]])\n \n dst=np.float32([[1058,20],\n [1058,700],\n [242,700],\n [242,20]])\n M = cv2.getPerspectiveTransform(src, dst)\n warped = cv2.warpPerspective(img, M, (1280,720), flags=cv2.INTER_LINEAR)\n \n return (warped, M)",
"_____no_output_____"
],
[
"def undistorT(imgorg): # Calculate Undistortion coefficients\n\n\n nx =9\n ny = 6\n objpoints = []\n imgpoints = []\n objp=np.zeros((6*9,3),np.float32)\n objp[:,:2]=np.mgrid[0:6,0:9].T.reshape(-1,2)\n\n\n images=glob.glob('./camera_cal/calibration*.jpg')\n for fname in images: # find corner points and Make a list of calibration images\n img = cv2.imread(fname)\n # Convert to grayscale\n gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\n # Find the chessboard corners\n ret, corners = cv2.findChessboardCorners(gray, (6,9),None)\n\n # If found, draw corners\n if ret == True:\n imgpoints.append(corners)\n objpoints.append(objp)\n # Draw and display the corners\n #cv2.drawChessboardCorners(img, (nx, ny), corners, ret) \n\n\n return cv2.calibrateCamera(objpoints,imgpoints,gray.shape[::-1],None,None)\n\n\n ",
"_____no_output_____"
],
[
"def undistresult(img, mtx,dist): # undistort frame\n undist= cv2.undistort(img, mtx, dist, None, mtx)\n \n return undist\n\n",
"_____no_output_____"
],
[
"def LineFitting(wimgun): #Fit Lane Lines\n\n # Set minimum number of pixels found to recenter window\n minpix = 20\n # Create empty lists to receive left and right lane pixel indices\n left_lane_inds = []\n right_lane_inds = []\n \n\n histogram = np.sum(wimgun[350:,:], axis=0)\n # Create an output image to draw on and visualize the result\n out_img = np.dstack((wimgun, wimgun, wimgun))\n\n\n # Find the peak of the left and right halves of the histogram\n # These will be the starting point for the left and right lines\n midpoint = np.int(histogram.shape[0]/2)\n leftx_base = np.argmax(histogram[:midpoint])\n\n rightx_base = np.argmax(histogram[midpoint:]) + midpoint\n nwindows = 9\n\n # Set height of windows\n window_height = np.int(wimgun.shape[0]/nwindows)\n # Identify the x and y positions of all nonzero pixels in the image\n nonzero = wimgun.nonzero()\n nonzeroy = np.array(nonzero[0])\n nonzerox = np.array(nonzero[1])\n # Current positions to be updated for each window\n leftx_current = leftx_base\n rightx_current = rightx_base\n # Set the width of the windows +/- margin\n margin =80\n\n\n\n\n # Step through the windows one by one\n for window in range(nwindows):\n # Identify window boundaries in x and y (and right and left)\n win_y_low = wimgun.shape[0] - (window+1)*window_height\n win_y_high = wimgun.shape[0] - window*window_height\n win_xleft_low = leftx_current - margin\n win_xleft_high = leftx_current + margin\n win_xright_low = rightx_current - margin\n win_xright_high = rightx_current + margin\n # Draw the windows on the visualization image\n cv2.rectangle(out_img,(win_xleft_low,win_y_low),(win_xleft_high,win_y_high),(0,255,0), 2) \n cv2.rectangle(out_img,(win_xright_low,win_y_low),(win_xright_high,win_y_high),(0,255,0), 2) \n # Identify the nonzero pixels in x and y within the window\n good_left_inds = ((nonzeroy >= win_y_low) & (nonzeroy < win_y_high) & (nonzerox >= win_xleft_low) & (nonzerox < win_xleft_high)).nonzero()[0]\n good_right_inds = ((nonzeroy >= win_y_low) & (nonzeroy < win_y_high) & (nonzerox >= win_xright_low) & (nonzerox < win_xright_high)).nonzero()[0]\n # Append these indices to the lists\n\n left_lane_inds.append(good_left_inds)\n right_lane_inds.append(good_right_inds)\n # If you found > minpix pixels, recenter next window on their mean position\n if len(good_left_inds) > minpix:\n leftx_current = np.int(np.mean(nonzerox[good_left_inds]))\n if len(good_right_inds) > minpix: \n rightx_current = np.int(np.mean(nonzerox[good_right_inds]))\n\n\n # Concatenate the arrays of indices\n left_lane_inds = np.concatenate(left_lane_inds)\n right_lane_inds = np.concatenate(right_lane_inds)\n # Again, extract left and right line pixel positions\n leftx = nonzerox[left_lane_inds]\n lefty = nonzeroy[left_lane_inds] \n rightx = nonzerox[right_lane_inds]\n righty = nonzeroy[right_lane_inds]\n\n\n # Fit a second order polynomial to each\n left_fit = np.polyfit(lefty, leftx, 2)\n right_fit = np.polyfit(righty, rightx, 2)\n # Generate x and y values for plotting\n ploty = np.linspace(0, wimgun.shape[0]-1, wimgun.shape[0] )\n left_fitx = left_fit[0]*ploty**2 + left_fit[1]*ploty + left_fit[2]\n right_fitx = right_fit[0]*ploty**2 + right_fit[1]*ploty + right_fit[2]\n\n # Create an image to draw on and an image to show the selection window\n# out_img = np.dstack((wimgun, wimgun, wimgun))*255\n window_img = np.zeros_like(out_img)\n # Color in left and right line pixels\n out_img[nonzeroy[left_lane_inds], nonzerox[left_lane_inds]] = [255, 0, 0]\n out_img[nonzeroy[right_lane_inds], nonzerox[right_lane_inds]] = [0, 0, 255]\n \n \n# plt.plot(left_fitx, ploty, color='yellow')\n# plt.plot(right_fitx, ploty, color='yellow')\n# plt.xlim(0, 1280)\n# plt.ylim(720, 0)\n# plt.imshow(out_img)\n# # plt.savefig(\"./output_images/Window Image\"+str(n)+\".png\")\n# plt.show()\n\n # Generate a polygon to illustrate the search window area\n # And recast the x and y points into usable format for cv2.fillPoly()\n left_line_window1 = np.array([np.transpose(np.vstack([left_fitx-margin, ploty]))])\n left_line_window2 = np.array([np.flipud(np.transpose(np.vstack([left_fitx+margin, ploty])))])\n left_line_pts = np.hstack((left_line_window1, left_line_window2))\n right_line_window1 = np.array([np.transpose(np.vstack([right_fitx-margin, ploty]))])\n right_line_window2 = np.array([np.flipud(np.transpose(np.vstack([right_fitx+margin, ploty])))])\n right_line_pts = np.hstack((right_line_window1, right_line_window2))\n\n # Draw the lane onto the warped blank image\n cv2.fillPoly(window_img, np.int_([left_line_pts]), (0,255, 0))\n cv2.fillPoly(window_img, np.int_([right_line_pts]), (0,255, 0))\n result = cv2.addWeighted(out_img, 1, window_img, 0.3, 0)\n# plt.title(\"r\")\n\n# plt.plot(left_fitx, ploty, color='yellow')\n# plt.plot(right_fitx, ploty, color='yellow')\n# plt.xlim(0, 1280)\n# plt.ylim(720, 0)\n# plt.imshow(result)\n# # plt.savefig(\"./output_images/Line Image\"+str(n)+\".png\")\n# plt.show()\n\n\n # Define y-value where we want radius of curvature\n # I'll choose the maximum y-value, corresponding to the bottom of the image\n y_eval = np.max(ploty)\n left_curverad = ((1 + (2*left_fit[0]*y_eval + left_fit[1])**2)**1.5) / np.absolute(2*left_fit[0])\n right_curverad = ((1 + (2*right_fit[0]*y_eval + right_fit[1])**2)**1.5) / np.absolute(2*right_fit[0])\n #print(left_curverad, right_curverad)\n \n ym_per_pix = 30/720 # meters per pixel in y dimension\n xm_per_pix = 3.7/700 # meters per pixel in x dimension\n\n# Fit new polynomials to x,y in world space\n left_fit_cr = np.polyfit(ploty*ym_per_pix, left_fitx*xm_per_pix, 2)\n right_fit_cr = np.polyfit(ploty*ym_per_pix, right_fitx*xm_per_pix, 2)\n# y_eval = np.max(ploty)\n\n# # Calculate the new radias of curvature\n \n left_curverad = ((1 + (2*left_fit_cr[0]*y_eval*ym_per_pix + left_fit_cr[1])**2)**1.5) / np.absolute(2*left_fit_cr[0])\n right_curverad = ((1 + (2*right_fit_cr[0]*y_eval*ym_per_pix + right_fit_cr[1])**2)**1.5) / np.absolute(2*right_fit_cr[0])\n# # left_curverad = ((1 + (2*left_fit[0]*y_eval + left_fit[1])**2)**1.5) / np.absolute(2*left_fit[0])\n# # right_curverad = ((1 + (2*right_fit[0]*y_eval + right_fit[1])**2)**1.5) / np.absolute(2*right_fit[0])\n \n \n# camera_center=wimgun.shape[0]/2\n# #lane_center = (right_fitx[719] + left_fitx[719])/2 \n lane_offset = (1280/2 - (left_fitx[-1]+right_fitx[-1])/2)*xm_per_pix\n# print(left_curverad1, right_curverad1, lane_offset)\n\n return (left_fit, ploty,right_fit,left_curverad, right_curverad,lane_offset)\n\n \n\n # Create an image to draw the lines on\ndef unwrappedframe(img,pm, Minv, left_fit,ploty,right_fit):\n left_fitx = left_fit[0]*ploty**2 + left_fit[1]*ploty + left_fit[2]\n right_fitx = right_fit[0]*ploty**2 + right_fit[1]*ploty + right_fit[2]\n nonzero = img.nonzero()\n nonzeroy = np.array(nonzero[0])\n nonzerox = np.array(nonzero[1])\n \n \n warp_zero = np.zeros_like(pm).astype(np.uint8)\n color_warp = np.dstack((warp_zero, warp_zero, warp_zero))\n\n # Recast the x and y points into usable format for cv2.fillPoly()\n pts_left = np.array([np.transpose(np.vstack([left_fitx, ploty]))])\n pts_right = np.array([np.flipud(np.transpose(np.vstack([right_fitx, ploty])))])\n pts = np.hstack((pts_left, pts_right))\n\n # Draw the lane onto the warped blank image\n cv2.fillPoly(color_warp, np.int_([pts]), (0,255, 0))\n\n # Warp the blank back to original image space using inverse perspective matrix (Minv)\n newwarp = cv2.warpPerspective(color_warp, Minv, (img.shape[1], img.shape[0])) \n \n # Combine the result with the original image\n\n return cv2.addWeighted(img, 1, newwarp, 0.3, 0)\n \n \n ",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0026c3e195defcec74621cf3f60c1e4f5892723 | 7,576 | ipynb | Jupyter Notebook | lect13_NumPy/2021_DPO_13_2_heroku.ipynb | weqrwer/Python_DPO_2021_fall | 8558ed2c1a744638f693ad036cfafccd1a05f392 | [
"MIT"
] | 3 | 2022-02-19T17:20:33.000Z | 2022-03-02T11:35:56.000Z | lect13_NumPy/2021_DPO_13_2_heroku.ipynb | weqrwer/Python_DPO_2021_fall | 8558ed2c1a744638f693ad036cfafccd1a05f392 | [
"MIT"
] | null | null | null | lect13_NumPy/2021_DPO_13_2_heroku.ipynb | weqrwer/Python_DPO_2021_fall | 8558ed2c1a744638f693ad036cfafccd1a05f392 | [
"MIT"
] | 8 | 2021-09-16T10:28:30.000Z | 2021-11-24T06:20:09.000Z | 25.0033 | 135 | 0.556494 | [
[
[
"## Как выложить бота на HEROKU\n\n*Подготовил Ян Пиле*",
"_____no_output_____"
],
[
"Сразу оговоримся, что мы на heroku выкладываем\n\n**echo-Бота в телеграме, написанного с помощью библиотеки [pyTelegramBotAPI](https://github.com/eternnoir/pyTelegramBotAPI)**.\n\nА взаимодействие его с сервером мы сделаем с использованием [flask](http://flask.pocoo.org/)\n\nТо есть вы боту пишете что-то, а он вам отвечает то же самое.",
"_____no_output_____"
],
[
"## Регистрация",
"_____no_output_____"
],
[
"Идем к **@BotFather** в Telegram и по его инструкции создаем нового бота командой **/newbot**. \n\nЭто должно закончиться выдачей вам токена вашего бота. Например последовательность команд, введенных мной:\n\n* **/newbot**\n* **my_echo_bot** (имя бота)\n* **ian_echo_bot** (ник бота в телеграме)\n\nЗавершилась выдачей мне токена **1403467808:AAEaaLPkIqrhrQ62p7ToJclLtNNINdOopYk**\n\nИ ссылки t.me/ian_echo_bot\n\n<img src=\"botfather.png\">",
"_____no_output_____"
],
[
"## Регистрация на HEROKU\n \nИдем сюда: https://signup.heroku.com/login\n\nСоздаем пользователя (это бесплатно)\n\nПопадаем на https://dashboard.heroku.com/apps и там создаем новое приложение:\n<img src=\"newapp1.png\">",
"_____no_output_____"
],
[
"Вводим название и регион (Я выбрал Европу), создаем.\n<img src=\"newapp2.png\">\n\nПосле того, как приложение создано, нажмите, \"Open App\" и скопируйте адрес оттуда. \n\n<img src=\"newapp3.png\">\n\nУ меня это https://ian-echo-bot.herokuapp.com",
"_____no_output_____"
],
[
"## Установить интерфейсы heroku и git для командной строки",
"_____no_output_____"
],
[
"Теперь надо установить Интерфейсы командной строки heroku и git по ссылкам:\n\n* https://devcenter.heroku.com/articles/heroku-cli\n* https://git-scm.com/book/en/v2/Getting-Started-Installing-Git",
"_____no_output_____"
],
[
"## Установить библиотеки",
"_____no_output_____"
],
[
"Теперь в вашем редакторе (например PyCharm) надо установить библиотеку для Телеграма и flask:\n\n* pip install pyTelegramBotAPI\n* pip install flask",
"_____no_output_____"
],
[
"## Код нашего echo-бота\n\nВот этот код я уложил в файл main.py",
"_____no_output_____"
]
],
[
[
"import os\nimport telebot\nfrom flask import Flask, request\n\nTOKEN = '1403467808:AAEaaLPkIqrhrQ62p7ToJclLtNNINdOopYk' # это мой токен\nbot = telebot.TeleBot(token=TOKEN)\nserver = Flask(__name__)\n\n \n# Если строка на входе непустая, то бот повторит ее\[email protected]_handler(func=lambda msg: msg.text is not None)\ndef reply_to_message(message):\n bot.send_message(message.chat.id, message.text)\n\[email protected]('/' + TOKEN, methods=['POST'])\ndef getMessage():\n bot.process_new_updates([telebot.types.Update.de_json(request.stream.read().decode(\"utf-8\"))])\n return \"!\", 200\n\[email protected](\"/\")\ndef webhook():\n bot.remove_webhook()\n bot.set_webhook(url='https://ian-echo-bot.herokuapp.com/' + TOKEN) #\n return \"!\", 200\n\nif __name__ == \"__main__\":\n server.run(host=\"0.0.0.0\", port=int(os.environ.get('PORT', 5000)))\n",
"_____no_output_____"
]
],
[
[
"## Теперь создаем еще два файла для запуска",
"_____no_output_____"
],
[
"**Procfile**(файл без расширения). Его надо открыть текстовым редактором и вписать туда строку:\n\n web: python main.py",
"_____no_output_____"
],
[
"**requirements.txt** - файл со списком версий необходимых библиотек. \n\nЗайдите в PyCharm, где вы делаете проект и введите в терминале команду:\n\n pip list format freeze > requirements.txt\n\nВ файле записи должны иметь вид:\n \n Название библиотеки Версия библиотеки\n \nЕсли вдруг вы выдите что-то такое:\n<img src=\"versions.png\">\n\nУдалите этот кусок текста, чтоб остался только номер версии и сохраните файл.\n\nТеперь надо все эти файлы уложить на гит, привязанный к Heroku и запустить приложение.\n\n",
"_____no_output_____"
],
[
"## Последний шаг",
"_____no_output_____"
],
[
"Надо залогиниться в heroku через командную строку.\n\nВведите:\n\n heroku login\n\nВас перебросит в браузер на вот такую страницу:\n<img src=\"login.png\">\n",
"_____no_output_____"
],
[
"После того, как вы залогинились, удостоверьтесь, что вы находитесь в папке, где лежат фаши файлы:\n \n main.py\n Procfile\n requirements.txt",
"_____no_output_____"
],
[
"**Вводите команды:**",
"_____no_output_____"
],
[
" git init \n git add .\n git commit -m \"first commit\"\n heroku git:remote -a ian-echo-bot\n git push heroku master",
"_____no_output_____"
],
[
"По ходу выкатки вы увидите что-то такое:\n<img src=\"process.png\">\n ",
"_____no_output_____"
],
[
"Готово, вы выложили вашего бота.",
"_____no_output_____"
],
[
"Материалы, которыми можно воспользоваться по ходу выкладки бота на сервер:\n\nhttps://towardsdatascience.com/how-to-deploy-a-telegram-bot-using-heroku-for-free-9436f89575d2\n\nhttps://mattrighetti.medium.com/build-your-first-telegram-bot-using-python-and-heroku-79d48950d4b0",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d00271ef6c25cb9f8943f0f3640f0c6209e44e85 | 29,784 | ipynb | Jupyter Notebook | Python for Finance - Code Files/83 Computing Alpha, Beta, and R Squared in Python/Python 2/Computing Alpha, Beta, and R Squared in Python - Solution.ipynb | siddharthjain1611/Python_for_Finance_Investment_Fundamentals-and-Data-Analytics | f2f1e22f2d578c59f833f8f3c8b4523d91286e9e | [
"MIT"
] | 3 | 2020-03-24T12:58:37.000Z | 2020-08-03T17:22:35.000Z | Python for Finance - Code Files/83 Computing Alpha, Beta, and R Squared in Python/Python 2/Computing Alpha, Beta, and R Squared in Python - Solution.ipynb | siddharthjain1611/Python_for_Finance_Investment_Fundamentals-and-Data-Analytics | f2f1e22f2d578c59f833f8f3c8b4523d91286e9e | [
"MIT"
] | null | null | null | Python for Finance - Code Files/83 Computing Alpha, Beta, and R Squared in Python/Python 2/Computing Alpha, Beta, and R Squared in Python - Solution.ipynb | siddharthjain1611/Python_for_Finance_Investment_Fundamentals-and-Data-Analytics | f2f1e22f2d578c59f833f8f3c8b4523d91286e9e | [
"MIT"
] | 1 | 2021-10-19T23:59:37.000Z | 2021-10-19T23:59:37.000Z | 58.514735 | 8,992 | 0.727001 | [
[
[
"## Computing Alpha, Beta, and R Squared in Python ",
"_____no_output_____"
],
[
"*Suggested Answers follow (usually there are multiple ways to solve a problem in Python).*",
"_____no_output_____"
],
[
"*Running a Regression in Python - continued:*",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom scipy import stats\nimport statsmodels.api as sm \nimport matplotlib.pyplot as plt\n\ndata = pd.read_excel('D:/Python/Data_Files/IQ_data.xlsx')\n\nX = data['Test 1']\nY = data['IQ']\n\nplt.scatter(X,Y)\nplt.axis([0, 120, 0, 150])\nplt.ylabel('IQ')\nplt.xlabel('Test 1')\nplt.show()",
"_____no_output_____"
]
],
[
[
"****",
"_____no_output_____"
],
[
"Use the statsmodels’ **.add_constant()** method to reassign the X data on X1. Use OLS with arguments Y and X1 and apply the fit method to obtain univariate regression results. Help yourself with the **.summary()** method. ",
"_____no_output_____"
]
],
[
[
"X1 = sm.add_constant(X)\n\nreg = sm.OLS(Y, X1).fit()",
"_____no_output_____"
],
[
"reg.summary()",
"_____no_output_____"
]
],
[
[
"By looking at the p-values, would you conclude Test 1 scores are a good predictor?",
"_____no_output_____"
],
[
"*****",
"_____no_output_____"
],
[
"Imagine a kid would score 84 on Test 1. How many points is she expected to get on the IQ test, approximately?",
"_____no_output_____"
]
],
[
[
"45 + 84*0.76",
"_____no_output_____"
]
],
[
[
"******",
"_____no_output_____"
],
[
"### Alpha, Beta, R^2:",
"_____no_output_____"
],
[
"Apply the stats module’s **linregress()** to extract the value for the slope, the intercept, the r squared, the p_value, and the standard deviation.",
"_____no_output_____"
]
],
[
[
"slope, intercept, r_value, p_value, std_err = stats.linregress(X,Y)",
"_____no_output_____"
],
[
"slope",
"_____no_output_____"
],
[
"intercept",
"_____no_output_____"
],
[
"r_value",
"_____no_output_____"
],
[
"r_value ** 2",
"_____no_output_____"
],
[
"p_value",
"_____no_output_____"
],
[
"std_err",
"_____no_output_____"
]
],
[
[
"Use the values of the slope and the intercept to predict the IQ score of a child, who obtained 84 points on Test 1. Is the forecasted value different than the one you obtained above?",
"_____no_output_____"
]
],
[
[
"intercept + 84 * slope",
"_____no_output_____"
]
],
[
[
"******",
"_____no_output_____"
],
[
"Follow the steps to draw the best fitting line of the provided regression.",
"_____no_output_____"
],
[
"Define a function that will use the slope and the intercept value to calculate the dots of the best fitting line.",
"_____no_output_____"
]
],
[
[
"def fitline(b):\n return intercept + slope * b",
"_____no_output_____"
]
],
[
[
"Apply it to the data you have stored in the variable X.",
"_____no_output_____"
]
],
[
[
"line = fitline(X)",
"_____no_output_____"
]
],
[
[
"Draw a scatter plot with the X and Y data and then plot X and the obtained fit-line.",
"_____no_output_____"
]
],
[
[
"plt.scatter(X,Y)\nplt.plot(X,line)\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0027ad3b4f0aacb9dd8f9d1e33562baa9f49a38 | 63,120 | ipynb | Jupyter Notebook | Kaggle_Challenge_Assignment_Submission5.ipynb | JimKing100/DS-Unit-2-Kaggle-Challenge | d1a705987c5a4df8b3ab74daab453754b77045cc | [
"MIT"
] | null | null | null | Kaggle_Challenge_Assignment_Submission5.ipynb | JimKing100/DS-Unit-2-Kaggle-Challenge | d1a705987c5a4df8b3ab74daab453754b77045cc | [
"MIT"
] | null | null | null | Kaggle_Challenge_Assignment_Submission5.ipynb | JimKing100/DS-Unit-2-Kaggle-Challenge | d1a705987c5a4df8b3ab74daab453754b77045cc | [
"MIT"
] | null | null | null | 50.821256 | 280 | 0.488926 | [
[
[
"<a href=\"https://colab.research.google.com/github/JimKing100/DS-Unit-2-Kaggle-Challenge/blob/master/Kaggle_Challenge_Assignment_Submission5.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# Installs\n%%capture\n!pip install --upgrade category_encoders plotly",
"_____no_output_____"
],
[
"# Imports\nimport os, sys\n\nos.chdir('/content')\n!git init .\n!git remote add origin https://github.com/LambdaSchool/DS-Unit-2-Kaggle-Challenge.git\n!git pull origin master\n\n!pip install -r requirements.txt\n\nos.chdir('module1')",
"Reinitialized existing Git repository in /content/.git/\nfatal: remote origin already exists.\nFrom https://github.com/LambdaSchool/DS-Unit-2-Kaggle-Challenge\n * branch master -> FETCH_HEAD\nAlready up to date.\nRequirement already satisfied: category_encoders==2.0.0 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 1)) (2.0.0)\nRequirement already satisfied: eli5==0.10.1 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 2)) (0.10.1)\nRequirement already satisfied: matplotlib!=3.1.1 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 3)) (3.0.3)\nRequirement already satisfied: pandas-profiling==2.3.0 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 4)) (2.3.0)\nRequirement already satisfied: pdpbox==0.2.0 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 5)) (0.2.0)\nRequirement already satisfied: plotly==4.1.1 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 6)) (4.1.1)\nRequirement already satisfied: seaborn==0.9.0 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 7)) (0.9.0)\nRequirement already satisfied: scikit-learn==0.21.3 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 8)) (0.21.3)\nRequirement already satisfied: shap==0.30.0 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 9)) (0.30.0)\nRequirement already satisfied: xgboost==0.90 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 10)) (0.90)\nRequirement already satisfied: scipy>=0.19.0 in /usr/local/lib/python3.6/dist-packages (from category_encoders==2.0.0->-r requirements.txt (line 1)) (1.3.1)\nRequirement already satisfied: statsmodels>=0.6.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders==2.0.0->-r requirements.txt (line 1)) (0.10.1)\nRequirement already satisfied: patsy>=0.4.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders==2.0.0->-r requirements.txt (line 1)) (0.5.1)\nRequirement already satisfied: numpy>=1.11.3 in /usr/local/lib/python3.6/dist-packages (from category_encoders==2.0.0->-r requirements.txt (line 1)) (1.16.5)\nRequirement already satisfied: pandas>=0.21.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders==2.0.0->-r requirements.txt (line 1)) (0.24.2)\nRequirement already satisfied: jinja2 in /usr/local/lib/python3.6/dist-packages (from eli5==0.10.1->-r requirements.txt (line 2)) (2.10.1)\nRequirement already satisfied: graphviz in /usr/local/lib/python3.6/dist-packages (from eli5==0.10.1->-r requirements.txt (line 2)) (0.10.1)\nRequirement already satisfied: tabulate>=0.7.7 in /usr/local/lib/python3.6/dist-packages (from eli5==0.10.1->-r requirements.txt (line 2)) (0.8.3)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from eli5==0.10.1->-r requirements.txt (line 2)) (1.12.0)\nRequirement already satisfied: attrs>16.0.0 in /usr/local/lib/python3.6/dist-packages (from eli5==0.10.1->-r requirements.txt (line 2)) (19.1.0)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.6/dist-packages (from matplotlib!=3.1.1->-r requirements.txt (line 3)) (0.10.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib!=3.1.1->-r requirements.txt (line 3)) (1.1.0)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib!=3.1.1->-r requirements.txt (line 3)) (2.4.2)\nRequirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib!=3.1.1->-r requirements.txt (line 3)) (2.5.3)\nRequirement already satisfied: confuse>=1.0.0 in /usr/local/lib/python3.6/dist-packages (from pandas-profiling==2.3.0->-r requirements.txt (line 4)) (1.0.0)\nRequirement already satisfied: missingno>=0.4.2 in /usr/local/lib/python3.6/dist-packages (from pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.4.2)\nRequirement already satisfied: astropy in /usr/local/lib/python3.6/dist-packages (from pandas-profiling==2.3.0->-r requirements.txt (line 4)) (3.0.5)\nRequirement already satisfied: htmlmin>=0.1.12 in /usr/local/lib/python3.6/dist-packages (from pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.1.12)\nRequirement already satisfied: phik>=0.9.8 in /usr/local/lib/python3.6/dist-packages (from pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.9.8)\nRequirement already satisfied: joblib in /usr/local/lib/python3.6/dist-packages (from pdpbox==0.2.0->-r requirements.txt (line 5)) (0.13.2)\nRequirement already satisfied: psutil in /usr/local/lib/python3.6/dist-packages (from pdpbox==0.2.0->-r requirements.txt (line 5)) (5.4.8)\nRequirement already satisfied: retrying>=1.3.3 in /usr/local/lib/python3.6/dist-packages (from plotly==4.1.1->-r requirements.txt (line 6)) (1.3.3)\nRequirement already satisfied: tqdm>4.25.0 in /usr/local/lib/python3.6/dist-packages (from shap==0.30.0->-r requirements.txt (line 9)) (4.28.1)\nRequirement already satisfied: scikit-image in /usr/local/lib/python3.6/dist-packages (from shap==0.30.0->-r requirements.txt (line 9)) (0.15.0)\nRequirement already satisfied: ipython in /usr/local/lib/python3.6/dist-packages (from shap==0.30.0->-r requirements.txt (line 9)) (5.5.0)\nRequirement already satisfied: pytz>=2011k in /usr/local/lib/python3.6/dist-packages (from pandas>=0.21.1->category_encoders==2.0.0->-r requirements.txt (line 1)) (2018.9)\nRequirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.6/dist-packages (from jinja2->eli5==0.10.1->-r requirements.txt (line 2)) (1.1.1)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from kiwisolver>=1.0.1->matplotlib!=3.1.1->-r requirements.txt (line 3)) (41.2.0)\nRequirement already satisfied: pyyaml in /usr/local/lib/python3.6/dist-packages (from confuse>=1.0.0->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (3.13)\nRequirement already satisfied: numba>=0.38.1 in /usr/local/lib/python3.6/dist-packages (from phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.40.1)\nRequirement already satisfied: pytest-pylint>=0.13.0 in /usr/local/lib/python3.6/dist-packages (from phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.14.1)\nRequirement already satisfied: pytest>=4.0.2 in /usr/local/lib/python3.6/dist-packages (from phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (5.1.2)\nRequirement already satisfied: jupyter-client>=5.2.3 in /usr/local/lib/python3.6/dist-packages (from phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (5.3.1)\nRequirement already satisfied: nbconvert>=5.3.1 in /usr/local/lib/python3.6/dist-packages (from phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (5.6.0)\nRequirement already satisfied: networkx>=2.0 in /usr/local/lib/python3.6/dist-packages (from scikit-image->shap==0.30.0->-r requirements.txt (line 9)) (2.3)\nRequirement already satisfied: imageio>=2.0.1 in /usr/local/lib/python3.6/dist-packages (from scikit-image->shap==0.30.0->-r requirements.txt (line 9)) (2.4.1)\nRequirement already satisfied: pillow>=4.3.0 in /usr/local/lib/python3.6/dist-packages (from scikit-image->shap==0.30.0->-r requirements.txt (line 9)) (4.3.0)\nRequirement already satisfied: PyWavelets>=0.4.0 in /usr/local/lib/python3.6/dist-packages (from scikit-image->shap==0.30.0->-r requirements.txt (line 9)) (1.0.3)\nRequirement already satisfied: simplegeneric>0.8 in /usr/local/lib/python3.6/dist-packages (from ipython->shap==0.30.0->-r requirements.txt (line 9)) (0.8.1)\nRequirement already satisfied: traitlets>=4.2 in /usr/local/lib/python3.6/dist-packages (from ipython->shap==0.30.0->-r requirements.txt (line 9)) (4.3.2)\nRequirement already satisfied: pygments in /usr/local/lib/python3.6/dist-packages (from ipython->shap==0.30.0->-r requirements.txt (line 9)) (2.1.3)\nRequirement already satisfied: pickleshare in /usr/local/lib/python3.6/dist-packages (from ipython->shap==0.30.0->-r requirements.txt (line 9)) (0.7.5)\nRequirement already satisfied: pexpect; sys_platform != \"win32\" in /usr/local/lib/python3.6/dist-packages (from ipython->shap==0.30.0->-r requirements.txt (line 9)) (4.7.0)\nRequirement already satisfied: decorator in /usr/local/lib/python3.6/dist-packages (from ipython->shap==0.30.0->-r requirements.txt (line 9)) (4.4.0)\nRequirement already satisfied: prompt-toolkit<2.0.0,>=1.0.4 in /usr/local/lib/python3.6/dist-packages (from ipython->shap==0.30.0->-r requirements.txt (line 9)) (1.0.16)\nRequirement already satisfied: llvmlite>=0.25.0dev0 in /usr/local/lib/python3.6/dist-packages (from numba>=0.38.1->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.29.0)\nRequirement already satisfied: pylint>=1.4.5 in /usr/local/lib/python3.6/dist-packages (from pytest-pylint>=0.13.0->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (2.3.1)\nRequirement already satisfied: importlib-metadata>=0.12; python_version < \"3.8\" in /usr/local/lib/python3.6/dist-packages (from pytest>=4.0.2->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.23)\nRequirement already satisfied: wcwidth in /usr/local/lib/python3.6/dist-packages (from pytest>=4.0.2->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.1.7)\nRequirement already satisfied: py>=1.5.0 in /usr/local/lib/python3.6/dist-packages (from pytest>=4.0.2->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (1.8.0)\nRequirement already satisfied: atomicwrites>=1.0 in /usr/local/lib/python3.6/dist-packages (from pytest>=4.0.2->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (1.3.0)\nRequirement already satisfied: pluggy<1.0,>=0.12 in /usr/local/lib/python3.6/dist-packages (from pytest>=4.0.2->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.13.0)\nRequirement already satisfied: more-itertools>=4.0.0 in /usr/local/lib/python3.6/dist-packages (from pytest>=4.0.2->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (7.2.0)\nRequirement already satisfied: packaging in /usr/local/lib/python3.6/dist-packages (from pytest>=4.0.2->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (19.1)\nRequirement already satisfied: tornado>=4.1 in /usr/local/lib/python3.6/dist-packages (from jupyter-client>=5.2.3->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (4.5.3)\nRequirement already satisfied: pyzmq>=13 in /usr/local/lib/python3.6/dist-packages (from jupyter-client>=5.2.3->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (17.0.0)\nRequirement already satisfied: jupyter-core in /usr/local/lib/python3.6/dist-packages (from jupyter-client>=5.2.3->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (4.5.0)\nRequirement already satisfied: defusedxml in /usr/local/lib/python3.6/dist-packages (from nbconvert>=5.3.1->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.6.0)\nRequirement already satisfied: bleach in /usr/local/lib/python3.6/dist-packages (from nbconvert>=5.3.1->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (3.1.0)\nRequirement already satisfied: testpath in /usr/local/lib/python3.6/dist-packages (from nbconvert>=5.3.1->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.4.2)\nRequirement already satisfied: nbformat>=4.4 in /usr/local/lib/python3.6/dist-packages (from nbconvert>=5.3.1->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (4.4.0)\nRequirement already satisfied: entrypoints>=0.2.2 in /usr/local/lib/python3.6/dist-packages (from nbconvert>=5.3.1->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.3)\nRequirement already satisfied: pandocfilters>=1.4.1 in /usr/local/lib/python3.6/dist-packages (from nbconvert>=5.3.1->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (1.4.2)\nRequirement already satisfied: mistune<2,>=0.8.1 in /usr/local/lib/python3.6/dist-packages (from nbconvert>=5.3.1->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.8.4)\nRequirement already satisfied: olefile in /usr/local/lib/python3.6/dist-packages (from pillow>=4.3.0->scikit-image->shap==0.30.0->-r requirements.txt (line 9)) (0.46)\nRequirement already satisfied: ipython-genutils in /usr/local/lib/python3.6/dist-packages (from traitlets>=4.2->ipython->shap==0.30.0->-r requirements.txt (line 9)) (0.2.0)\nRequirement already satisfied: ptyprocess>=0.5 in /usr/local/lib/python3.6/dist-packages (from pexpect; sys_platform != \"win32\"->ipython->shap==0.30.0->-r requirements.txt (line 9)) (0.6.0)\nRequirement already satisfied: isort<5,>=4.2.5 in /usr/local/lib/python3.6/dist-packages (from pylint>=1.4.5->pytest-pylint>=0.13.0->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (4.3.21)\nRequirement already satisfied: astroid<3,>=2.2.0 in /usr/local/lib/python3.6/dist-packages (from pylint>=1.4.5->pytest-pylint>=0.13.0->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (2.2.5)\nRequirement already satisfied: mccabe<0.7,>=0.6 in /usr/local/lib/python3.6/dist-packages (from pylint>=1.4.5->pytest-pylint>=0.13.0->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.6.1)\nRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.6/dist-packages (from importlib-metadata>=0.12; python_version < \"3.8\"->pytest>=4.0.2->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.6.0)\nRequirement already satisfied: webencodings in /usr/local/lib/python3.6/dist-packages (from bleach->nbconvert>=5.3.1->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (0.5.1)\nRequirement already satisfied: jsonschema!=2.5.0,>=2.4 in /usr/local/lib/python3.6/dist-packages (from nbformat>=4.4->nbconvert>=5.3.1->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (2.6.0)\nRequirement already satisfied: lazy-object-proxy in /usr/local/lib/python3.6/dist-packages (from astroid<3,>=2.2.0->pylint>=1.4.5->pytest-pylint>=0.13.0->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (1.4.2)\nRequirement already satisfied: wrapt in /usr/local/lib/python3.6/dist-packages (from astroid<3,>=2.2.0->pylint>=1.4.5->pytest-pylint>=0.13.0->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (1.11.2)\nRequirement already satisfied: typed-ast>=1.3.0; implementation_name == \"cpython\" in /usr/local/lib/python3.6/dist-packages (from astroid<3,>=2.2.0->pylint>=1.4.5->pytest-pylint>=0.13.0->phik>=0.9.8->pandas-profiling==2.3.0->-r requirements.txt (line 4)) (1.4.0)\n"
],
[
"# Disable warning\nimport warnings\nwarnings.filterwarnings(action='ignore', category=FutureWarning, module='numpy')",
"_____no_output_____"
],
[
"# Imports\nimport pandas as pd\nimport numpy as np\nimport math\n\nimport sklearn\nsklearn.__version__\n\n# Import the models\nfrom sklearn.linear_model import LogisticRegressionCV\nfrom sklearn.pipeline import make_pipeline\n\n# Import encoder and scaler and imputer\nimport category_encoders as ce\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.impute import SimpleImputer\n\n# Import random forest classifier\nfrom sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"# Import, load data and split data into train, validate and test\ntrain_features = pd.read_csv('../data/tanzania/train_features.csv')\ntrain_labels = pd.read_csv('../data/tanzania/train_labels.csv')\ntest_features = pd.read_csv('../data/tanzania/test_features.csv')\nsample_submission = pd.read_csv('../data/tanzania/sample_submission.csv')\n\nassert train_features.shape == (59400, 40)\nassert train_labels.shape == (59400, 2)\nassert test_features.shape == (14358, 40)\nassert sample_submission.shape == (14358, 2)\n\n# Load initial train features and labels\nfrom sklearn.model_selection import train_test_split\nX_train = train_features\ny_train = train_labels['status_group']\n\n# Split the initial train features and labels 80% into new train and new validation\nX_train, X_val, y_train, y_val = train_test_split(\n X_train, y_train, train_size = 0.80, test_size = 0.20,\n stratify = y_train, random_state=42\n)\n\nX_train.shape, X_val.shape, y_train.shape, y_val.shape",
"_____no_output_____"
],
[
"# Wrangle train, validate, and test sets\ndef wrangle(X):\n \n # Set bins value\n bins=20\n chars = 3\n \n # Prevent SettingWithCopyWarning\n X = X.copy()\n \n X['latitude'] = X['latitude'].replace(-2e-08, 0)\n \n # Create missing columns\n cols_with_zeros = ['longitude', 'latitude', 'construction_year', \n 'gps_height', 'population']\n \n for col in cols_with_zeros:\n X[col] = X[col].replace(0, np.nan)\n X[col+'_missing'] = X[col].isnull()\n \n for col in cols_with_zeros:\n X[col] = X[col].replace(np.nan, 0)\n \n # Clean installer\n X['installer'] = X['installer'].str.lower()\n X['installer'] = X['installer'].str.replace('danid', 'danida')\n X['installer'] = X['installer'].str.replace('disti', 'district council')\n X['installer'] = X['installer'].str.replace('commu', 'community')\n X['installer'] = X['installer'].str.replace('central government', 'government')\n X['installer'] = X['installer'].str.replace('kkkt _ konde and dwe', 'kkkt')\n X['installer'] = X['installer'].str[:chars]\n X['installer'].value_counts(normalize=True)\n tops = X['installer'].value_counts()[:5].index\n X.loc[~X['installer'].isin(tops), 'installer'] = 'Other'\n \n # Clean funder and bin\n X['funder'] = X['funder'].str.lower()\n X['funder'] = X['funder'].str[:chars]\n X['funder'].value_counts(normalize=True)\n tops = X['funder'].value_counts()[:20].index\n X.loc[~X['funder'].isin(tops), 'funder'] = 'Other'\n\n # Use mean for gps_height missing values\n X.loc[X['gps_height'] == 0, 'gps_height'] = X['gps_height'].mean()\n \n # Bin lga\n tops = X['lga'].value_counts()[:10].index\n X.loc[~X['lga'].isin(tops), 'lga'] = 'Other'\n\n # Bin ward \n tops = X['ward'].value_counts()[:20].index\n X.loc[~X['ward'].isin(tops), 'ward'] = 'Other'\n \n # Bin subvillage\n tops = X['subvillage'].value_counts()[:bins].index\n X.loc[~X['subvillage'].isin(tops), 'subvillage'] = 'Other'\n\n # Clean latitude and longitude\n avg_lat_ward = X.groupby('ward').latitude.mean()\n avg_lat_lga = X.groupby('lga').latitude.mean()\n avg_lat_region = X.groupby('region').latitude.mean()\n avg_lat_country = X.latitude.mean()\n \n avg_long_ward = X.groupby('ward').longitude.mean()\n avg_long_lga = X.groupby('lga').longitude.mean()\n avg_long_region = X.groupby('region').longitude.mean()\n avg_long_country = X.longitude.mean()\n \n \n #cols_with_zeros = ['longitude', 'latitude']\n #for col in cols_with_zeros:\n # X[col] = X[col].replace(0, np.nan)\n #X.loc[X['latitude'] == 0, 'latitude'] = X['latitude'].median()\n #X.loc[X['longitude'] == 0, 'longitude'] = X['longitude'].median()\n \n #for i in range(0, 9): \n \n # X.loc[(X['latitude'] == 0) & (X['ward'] == avg_lat_ward.index[0]), 'latitude'] = avg_lat_ward[i]\n # X.loc[(X['latitude'] == 0) & (X['lga'] == avg_lat_lga.index[0]), 'latitude'] = avg_lat_lga[i]\n # X.loc[(X['latitude'] == 0) & (X['region'] == avg_lat_region.index[0]), 'latitude'] = avg_lat_region[i]\n # X.loc[(X['latitude'] == 0), 'latitude'] = avg_lat_country\n\n # X.loc[(X['longitude'] == 0) & (X['ward'] == avg_long_ward.index[0]), 'longitude'] = avg_long_ward[i]\n # X.loc[(X['longitude'] == 0) & (X['lga'] == avg_long_lga.index[0]), 'longitude'] = avg_long_lga[i]\n # X.loc[(X['longitude'] == 0) & (X['region'] == avg_long_region.index[0]), 'longitude'] = avg_long_region[i]\n # X.loc[(X['longitude'] == 0), 'longitude'] = avg_long_country\n \n average_lat = X.groupby('region').latitude.mean().reset_index()\n average_long = X.groupby('region').longitude.mean().reset_index()\n\n shinyanga_lat = average_lat.loc[average_lat['region'] == 'Shinyanga', 'latitude']\n shinyanga_long = average_long.loc[average_lat['region'] == 'Shinyanga', 'longitude']\n\n X.loc[(X['region'] == 'Shinyanga') & (X['latitude'] > -1), ['latitude']] = shinyanga_lat[17]\n X.loc[(X['region'] == 'Shinyanga') & (X['longitude'] == 0), ['longitude']] = shinyanga_long[17]\n\n mwanza_lat = average_lat.loc[average_lat['region'] == 'Mwanza', 'latitude']\n mwanza_long = average_long.loc[average_lat['region'] == 'Mwanza', 'longitude']\n\n X.loc[(X['region'] == 'Mwanza') & (X['latitude'] > -1), ['latitude']] = mwanza_lat[13]\n X.loc[(X['region'] == 'Mwanza') & (X['longitude'] == 0) , ['longitude']] = mwanza_long[13]\n \n # Impute mean for tsh based on mean of source_class/basin/waterpoint_type_group\n def tsh_calc(tsh, source, base, waterpoint):\n if tsh == 0:\n if (source, base, waterpoint) in tsh_dict:\n new_tsh = tsh_dict[source, base, waterpoint]\n return new_tsh\n else:\n return tsh\n return tsh\n \n temp = X[X['amount_tsh'] != 0].groupby(['source_class',\n 'basin',\n 'waterpoint_type_group'])['amount_tsh'].mean()\n\n tsh_dict = dict(temp)\n X['amount_tsh'] = X.apply(lambda x: tsh_calc(x['amount_tsh'], x['source_class'], x['basin'], x['waterpoint_type_group']), axis=1)\n X.loc[X['amount_tsh'] == 0, 'amount_tsh'] = X['amount_tsh'].median()\n \n # Impute mean for construction_year based on mean of source_class/basin/waterpoint_type_group\n #temp = X[X['construction_year'] != 0].groupby(['source_class',\n # 'basin',\n # 'waterpoint_type_group'])['amount_tsh'].mean()\n\n #tsh_dict = dict(temp)\n #X['construction_year'] = X.apply(lambda x: tsh_calc(x['construction_year'], x['source_class'], x['basin'], x['waterpoint_type_group']), axis=1)\n #X.loc[X['construction_year'] == 0, 'construction_year'] = X['construction_year'].mean()\n \n # Impute mean for the feature based on latitude and longitude\n def latlong_conversion(feature, pop, long, lat):\n \n radius = 0.1\n radius_increment = 0.3\n \n if pop <= 1:\n pop_temp = pop\n while pop_temp <= 1 and radius <= 2:\n lat_from = lat - radius\n lat_to = lat + radius\n long_from = long - radius\n long_to = long + radius\n \n df = X[(X['latitude'] >= lat_from) & \n (X['latitude'] <= lat_to) &\n (X['longitude'] >= long_from) &\n (X['longitude'] <= long_to)]\n \n pop_temp = df[feature].mean()\n if math.isnan(pop_temp):\n pop_temp = pop\n radius = radius + radius_increment\n else:\n pop_temp = pop\n \n if pop_temp <= 1:\n new_pop = X_train[feature].mean()\n else:\n new_pop = pop_temp\n \n return new_pop\n \n # Impute population based on location\n #X['population'] = X.apply(lambda x: latlong_conversion('population', x['population'], x['longitude'], x['latitude']), axis=1)\n #X.loc[X['population'] == 0, 'population'] = X['population'].median()\n \n # Impute gps_height based on location\n #X['gps_height'] = X.apply(lambda x: latlong_conversion('gps_height', x['gps_height'], x['longitude'], x['latitude']), axis=1)\n \n # Drop recorded_by (never varies) and id (always varies, random) and num_private (empty)\n unusable_variance = ['recorded_by', 'id', 'num_private','wpt_name', 'extraction_type_class',\n 'quality_group', 'source_type', 'source_class', 'waterpoint_type_group']\n X = X.drop(columns=unusable_variance)\n \n # Drop duplicate columns\n duplicates = ['quantity_group', 'payment_type', 'extraction_type_group']\n X = X.drop(columns=duplicates)\n \n # return the wrangled dataframe\n return X\n",
"_____no_output_____"
],
[
"# Wrangle the data\nX_train = wrangle(X_train)\nX_val = wrangle(X_val)",
"_____no_output_____"
],
[
"# Feature engineering\ndef feature_engineer(X):\n \n # Create new feature pump_age\n X['pump_age'] = 2013 - X['construction_year']\n X.loc[X['pump_age'] == 2013, 'pump_age'] = 0\n X.loc[X['pump_age'] == 0, 'pump_age'] = 10\n \n \n # Convert date_recorded to datetime\n X['date_recorded'] = pd.to_datetime(X['date_recorded'], infer_datetime_format=True)\n \n # Extract components from date_recorded, then drop the original column\n X['year_recorded'] = X['date_recorded'].dt.year\n X['month_recorded'] = X['date_recorded'].dt.month\n X['day_recorded'] = X['date_recorded'].dt.day\n \n # Engineer feature: how many years from construction_year to date_recorded\n X['years'] = X['year_recorded'] - X['construction_year']\n X['years_missing'] = X['years'].isnull()\n \n column_list = ['date_recorded']\n X = X.drop(columns=column_list)\n \n # Create new feature region_district\n X['region_district'] = X['region_code'].astype(str) + X['district_code'].astype(str)\n \n #X['tsh_pop'] = X['amount_tsh']/X['population']\n\n return X",
"_____no_output_____"
],
[
"# Feature engineer the data\nX_train = feature_engineer(X_train)\nX_val = feature_engineer(X_val)",
"_____no_output_____"
],
[
"X_train.head()",
"_____no_output_____"
],
[
"# Encode a feature\ndef encode_feature(X, y, str):\n X['status_group'] = y\n X.groupby(str)['status_group'].value_counts(normalize=True)\n X['functional']= (X['status_group'] == 'functional').astype(int)\n X[['status_group', 'functional']]\n return X",
"_____no_output_____"
],
[
"# Encode all the categorical features\ntrain = X_train.copy()\ntrain = encode_feature(train, y_train, 'quantity')\ntrain = encode_feature(train, y_train, 'waterpoint_type')\ntrain = encode_feature(train, y_train, 'extraction_type')\ntrain = encode_feature(train, y_train, 'installer')\ntrain = encode_feature(train, y_train, 'funder')\ntrain = encode_feature(train, y_train, 'water_quality')\ntrain = encode_feature(train, y_train, 'basin')\ntrain = encode_feature(train, y_train, 'region')\ntrain = encode_feature(train, y_train, 'payment')\ntrain = encode_feature(train, y_train, 'source')\ntrain = encode_feature(train, y_train, 'lga')\ntrain = encode_feature(train, y_train, 'ward')\ntrain = encode_feature(train, y_train, 'scheme_management')\ntrain = encode_feature(train, y_train, 'management')\ntrain = encode_feature(train, y_train, 'region_district')\ntrain = encode_feature(train, y_train, 'subvillage')",
"_____no_output_____"
],
[
"# use quantity feature and the numerical features but drop id\ncategorical_features = ['quantity', 'waterpoint_type', 'extraction_type', 'installer',\n 'basin', 'region', 'payment', 'source', 'lga', 'public_meeting',\n 'scheme_management', 'permit', 'management', 'region_district',\n 'subvillage', 'funder', 'water_quality', 'ward', 'years_missing', 'longitude_missing',\n 'latitude_missing','construction_year_missing', 'gps_height_missing',\n 'population_missing']\n \n# \nnumeric_features = X_train.select_dtypes('number').columns.tolist()\nfeatures = categorical_features + numeric_features\n\n# make subsets using the quantity feature all numeric features except id\nX_train = X_train[features]\nX_val = X_val[features]\n\n# Create the logistic regression pipeline\npipeline = make_pipeline (\n ce.OneHotEncoder(use_cat_names=True),\n #SimpleImputer(),\n StandardScaler(),\n LogisticRegressionCV(random_state=42, n_jobs=-1)\n)\n\npipeline.fit(X_train, y_train)\n\nprint('Validation Accuracy', pipeline.score(X_val, y_val)) ",
"/usr/local/lib/python3.6/dist-packages/sklearn/linear_model/logistic.py:469: FutureWarning: Default multi_class will be changed to 'auto' in 0.22. Specify the multi_class option to silence this warning.\n \"this warning.\", FutureWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/model_selection/_split.py:1978: FutureWarning: The default value of cv will change from 3 to 5 in version 0.22. Specify it explicitly to silence this warning.\n warnings.warn(CV_WARNING, FutureWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/linear_model/logistic.py:947: ConvergenceWarning: lbfgs failed to converge. Increase the number of iterations.\n \"of iterations.\", ConvergenceWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/linear_model/logistic.py:947: ConvergenceWarning: lbfgs failed to converge. Increase the number of iterations.\n \"of iterations.\", ConvergenceWarning)\n"
],
[
"features",
"_____no_output_____"
],
[
"# Create the random forest pipeline\npipeline = make_pipeline (\n ce.OrdinalEncoder(),\n SimpleImputer(strategy='mean'),\n StandardScaler(),\n RandomForestClassifier(n_estimators=1400, \n random_state=42,\n min_samples_split=5,\n min_samples_leaf=1,\n max_features='auto',\n max_depth=30,\n bootstrap=True,\n n_jobs=-1,\n verbose = 1)\n)\n\npipeline.fit(X_train, y_train)\nprint('Validation Accuracy', pipeline.score(X_val, y_val)) ",
"[Parallel(n_jobs=-1)]: Using backend ThreadingBackend with 4 concurrent workers.\n[Parallel(n_jobs=-1)]: Done 42 tasks | elapsed: 1.4s\n[Parallel(n_jobs=-1)]: Done 192 tasks | elapsed: 6.2s\n[Parallel(n_jobs=-1)]: Done 442 tasks | elapsed: 14.2s\n[Parallel(n_jobs=-1)]: Done 792 tasks | elapsed: 25.6s\n[Parallel(n_jobs=-1)]: Done 1242 tasks | elapsed: 40.5s\n[Parallel(n_jobs=-1)]: Done 1400 out of 1400 | elapsed: 45.7s finished\n[Parallel(n_jobs=4)]: Using backend ThreadingBackend with 4 concurrent workers.\n[Parallel(n_jobs=4)]: Done 42 tasks | elapsed: 0.1s\n[Parallel(n_jobs=4)]: Done 192 tasks | elapsed: 0.3s\n[Parallel(n_jobs=4)]: Done 442 tasks | elapsed: 0.6s\n[Parallel(n_jobs=4)]: Done 792 tasks | elapsed: 1.0s\n[Parallel(n_jobs=4)]: Done 1242 tasks | elapsed: 1.6s\n[Parallel(n_jobs=4)]: Done 1400 out of 1400 | elapsed: 1.8s finished\n"
],
[
"pd.set_option('display.max_rows', 200)\nmodel = pipeline.named_steps['randomforestclassifier']\nencoder = pipeline.named_steps['ordinalencoder']\nencoded_columns = encoder.transform(X_train).columns \nimportances = pd.Series(model.feature_importances_, encoded_columns)\nimportances.sort_values(ascending=False)",
"_____no_output_____"
],
[
"# Create missing columns\ncols_with_zeros = ['longitude', 'latitude', 'construction_year', \n 'gps_height', 'population']\n \nfor col in cols_with_zeros:\n test_features[col] = test_features[col].replace(0, np.nan)\n test_features[col+'_missing'] = test_features[col].isnull()\n \nfor col in cols_with_zeros:\n test_features[col] = test_features[col].replace(np.nan, 0)\n\ntest_features['pump_age'] = 2013 - test_features['construction_year']\ntest_features.loc[test_features['pump_age'] == 2013, 'pump_age'] = 0\ntest_features.loc[test_features['pump_age'] == 0, 'pump_age'] = 10\n \n# Convert date_recorded to datetime\ntest_features['date_recorded'] = pd.to_datetime(test_features['date_recorded'], infer_datetime_format=True)\n \n# Extract components from date_recorded, then drop the original column\ntest_features['year_recorded'] = test_features['date_recorded'].dt.year\ntest_features['month_recorded'] = test_features['date_recorded'].dt.month\ntest_features['day_recorded'] = test_features['date_recorded'].dt.day\n \n# Engineer feature: how many years from construction_year to date_recorded\ntest_features['years'] = test_features['year_recorded'] - test_features['construction_year']\ntest_features['years_missing'] = test_features['years'].isnull()\n\ntest_features['region_district'] = test_features['region_code'].astype(str) + test_features['district_code'].astype(str)\n \ncolumn_list = ['recorded_by', 'id', 'num_private','wpt_name', 'extraction_type_class',\n 'quality_group', 'source_type', 'source_class', 'waterpoint_type_group',\n 'quantity_group', 'payment_type', 'extraction_type_group']\n\ntest_features = test_features.drop(columns=column_list)\n\nX_test = test_features[features]\n\nassert all(X_test.columns == X_train.columns)\n\ny_pred = pipeline.predict(X_test)",
"[Parallel(n_jobs=4)]: Using backend ThreadingBackend with 4 concurrent workers.\n[Parallel(n_jobs=4)]: Done 42 tasks | elapsed: 0.1s\n[Parallel(n_jobs=4)]: Done 192 tasks | elapsed: 0.3s\n[Parallel(n_jobs=4)]: Done 442 tasks | elapsed: 0.6s\n[Parallel(n_jobs=4)]: Done 792 tasks | elapsed: 1.0s\n[Parallel(n_jobs=4)]: Done 1242 tasks | elapsed: 1.6s\n[Parallel(n_jobs=4)]: Done 1400 out of 1400 | elapsed: 1.8s finished\n"
],
[
"submission = sample_submission.copy()\nsubmission['status_group'] = y_pred\nsubmission.to_csv('/content/submission-05.csv', index=False)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0027f0c7d75bd8bb29a639ddc1b5f2c87a4cb6c | 2,897 | ipynb | Jupyter Notebook | Simple Interest, compound interest.ipynb | nankris/Puzzles-Questions- | 5044f84054e94ba9420c6239fd2170eed7007d00 | [
"Apache-2.0"
] | null | null | null | Simple Interest, compound interest.ipynb | nankris/Puzzles-Questions- | 5044f84054e94ba9420c6239fd2170eed7007d00 | [
"Apache-2.0"
] | null | null | null | Simple Interest, compound interest.ipynb | nankris/Puzzles-Questions- | 5044f84054e94ba9420c6239fd2170eed7007d00 | [
"Apache-2.0"
] | null | null | null | 17.993789 | 45 | 0.480152 | [
[
[
"#simple interest = ptr/100\n#p=principle amount\n#t is time (units of time)\n#r is rate (percent of interest)\np=int(input(\"principle amount\"))\nt=int(input(\"units of time\"))\nr=int(input(\"percent of interest\"))",
"principle amount10000\nunits of time5\npercent of interest5\n"
],
[
"simpleinterest=(p*t*r)/100",
"_____no_output_____"
],
[
"simpleinterest",
"_____no_output_____"
],
[
"#compound interest\n#CI=P(1+R/100)^r\n#P=Principle amount\n#R is rate\n#T is time span\n\np=int(input(\"principle amount\"))\nt=float(input(\"units of time\"))\nr=float(input(\"percent of interest\"))",
"principle amount1200\nunits of time2\npercent of interest5.4\n"
],
[
"import math\nci=p*pow((1+r/100),t)\nprint(ci)",
"1333.0992\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d00283e186c87119f70280c9f6b55acf172e7287 | 612,682 | ipynb | Jupyter Notebook | Combining-Thresholds.ipynb | joshrwhite/CarND-LaneLines-P1 | dbb2dbf9e3569b85c2613524dcedcaf5e3d54e84 | [
"MIT"
] | null | null | null | Combining-Thresholds.ipynb | joshrwhite/CarND-LaneLines-P1 | dbb2dbf9e3569b85c2613524dcedcaf5e3d54e84 | [
"MIT"
] | null | null | null | Combining-Thresholds.ipynb | joshrwhite/CarND-LaneLines-P1 | dbb2dbf9e3569b85c2613524dcedcaf5e3d54e84 | [
"MIT"
] | null | null | null | 4,505.014706 | 607,340 | 0.961143 | [
[
[
"import numpy as np\nimport cv2\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport pickle\n\n# Read in an image\nimage = mpimg.imread('signs_vehicles_xygrad.png')\n\ndef abs_sobel_thresh(img, orient='x', sobel_kernel=3, thresh=(0, 255)):\n # Apply the following steps to img\n # 1) Convert to grayscale\n gray = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)\n # 2) Take the derivative in x or y given orient = 'x' or 'y'\n sobelx = cv2.Sobel(gray, cv2.CV_64F, 1, 0)\n sobely = cv2.Sobel(gray, cv2.CV_64F, 0, 1)\n # 3) Take the absolute value of the derivative or gradient\n abs_sobelx = np.absolute(sobelx)\n # 4) Scale to 8-bit (0 - 255) then convert to type = np.uint8\n scaled_sobel = np.uint8(255*abs_sobelx/np.max(abs_sobelx))\n # 5) Create a mask of 1's where the scaled gradient magnitude\n # is > thresh_min and < thresh_max\n grad_binary = np.zeros_like(scaled_sobel)\n grad_binary[(scaled_sobel >= thresh[0]) & (scaled_sobel <= thresh[1])] = 1\n # 6) Return this mask as your binary_output image\n return grad_binary\n\ndef mag_thresh(image, sobel_kernel=3, mag_thresh=(0, 255)):\n # Apply the following steps to img\n # 1) Convert to grayscale\n gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)\n # 2) Take the derivative in x or y given orient = 'x' or 'y'\n sobelx = cv2.Sobel(gray, cv2.CV_64F, 1, 0, ksize=sobel_kernel)\n sobely = cv2.Sobel(gray, cv2.CV_64F, 0, 1, ksize=sobel_kernel)\n # 3) Calculate the magnitude \n abs_sobelx = np.sqrt(np.square(sobelx)+np.square(sobely))\n # 4) Scale to 8-bit (0 - 255) and convert to type = np.uint8\n scaled_sobel = np.uint8(255*abs_sobelx/np.max(abs_sobelx))\n # 5) Create a binary mask where mag thresholds are met\n mag_binary = np.zeros_like(scaled_sobel)\n mag_binary[(scaled_sobel >= mag_thresh[0]) & (scaled_sobel <= mag_thresh[1])] = 1\n # 6) Return this mask as your binary_output image\n return mag_binary\n\ndef dir_threshold(image, sobel_kernel=3, thresh=(0, np.pi/2)):\n # Apply the following steps to img\n # 1) Convert to grayscale\n gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)\n # 2) Take the derivative in x or y given orient = 'x' or 'y'\n sobelx = cv2.Sobel(gray, cv2.CV_64F, 1, 0, ksize=sobel_kernel)\n sobely = cv2.Sobel(gray, cv2.CV_64F, 0, 1, ksize=sobel_kernel)\n # 3) Take the absolute value of the x and y gradients\n abs_sobelx = np.absolute(sobelx)\n abs_sobely = np.absolute(sobely)\n # 4) Use np.arctan2(abs_sobely, abs_sobelx) to calculate the direction of the gradient \n grad_dir = np.arctan2(abs_sobely, abs_sobelx)\n # 5) Create a binary mask where direction thresholds are met\n dir_binary = np.zeros_like(grad_dir)\n dir_binary[(grad_dir >= thresh[0]) & (grad_dir <= thresh[1])] = 1\n # 6) Return this mask as your binary_output image\n return dir_binary\n\n# Choose a Sobel kernel size\nksize = 3 # Choose a larger odd number to smooth gradient measurements\n\n# Apply each of the thresholding functions\ngradx = abs_sobel_thresh(image, orient='x', sobel_kernel=ksize, thresh=(20, 100))\ngrady = abs_sobel_thresh(image, orient='y', sobel_kernel=ksize, thresh=(80, 100))\nmag_binary = mag_thresh(image, sobel_kernel=ksize, mag_thresh=(30, 100))\ndir_binary = dir_threshold(image, sobel_kernel=ksize, thresh=(0.7, 1.3))\n\ncombined = np.zeros_like(dir_binary)\ncombined[((gradx == 1) & (grady == 1)) | ((mag_binary == 1) & (dir_binary == 1))] = 1\n\n# Plot the result\nf, (ax1, ax2) = plt.subplots(1, 2, figsize=(24, 9))\nf.tight_layout()\nax1.imshow(image)\nax1.set_title('Original Image', fontsize=50)\nax2.imshow(combined, cmap='gray')\nax2.set_title('Combined Thresholds', fontsize=50)\nplt.subplots_adjust(left=0., right=1, top=0.9, bottom=0.)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d0029a295b258a0e9387ce34994b69142f1dd639 | 297,382 | ipynb | Jupyter Notebook | simulations.ipynb | diozaka/eitest | b2c37ad93e7760673a2f46279f913bd03440a8f2 | [
"MIT"
] | 2 | 2020-05-21T11:53:20.000Z | 2020-11-01T06:12:49.000Z | simulations.ipynb | diozaka/eitest | b2c37ad93e7760673a2f46279f913bd03440a8f2 | [
"MIT"
] | null | null | null | simulations.ipynb | diozaka/eitest | b2c37ad93e7760673a2f46279f913bd03440a8f2 | [
"MIT"
] | null | null | null | 219.30826 | 30,184 | 0.896709 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport numba\nfrom tqdm import tqdm\n\nimport eitest",
"_____no_output_____"
]
],
[
[
"# Data generators",
"_____no_output_____"
]
],
[
[
"@numba.njit\ndef event_series_bernoulli(series_length, event_count):\n '''Generate an iid Bernoulli distributed event series.\n\n series_length: length of the event series\n event_count: number of events'''\n\n event_series = np.zeros(series_length)\n event_series[np.random.choice(np.arange(0, series_length), event_count, replace=False)] = 1\n return event_series\n\[email protected]\ndef time_series_mean_impact(event_series, order, signal_to_noise):\n '''Generate a time series with impacts in mean as described in the paper.\n\n The impact weights are sampled iid from N(0, signal_to_noise),\n and additional noise is sampled iid from N(0,1). The detection problem will\n be harder than in time_series_meanconst_impact for small orders, as for small\n orders we have a low probability to sample at least one impact weight with a\n high magnitude. On the other hand, since the impact is different at every lag,\n we can detect the impacts even if the order is larger than the max_lag value\n used in the test.\n \n event_series: input of shape (T,) with event occurrences\n order: order of the event impacts\n signal_to_noise: signal to noise ratio of the event impacts'''\n\n series_length = len(event_series)\n weights = np.random.randn(order)*np.sqrt(signal_to_noise)\n time_series = np.random.randn(series_length)\n for t in range(series_length):\n if event_series[t] == 1:\n time_series[t+1:t+order+1] += weights[:order-max(0, (t+order+1)-series_length)]\n return time_series\n\[email protected]\ndef time_series_meanconst_impact(event_series, order, const):\n '''Generate a time series with impacts in mean by adding a constant.\n Better for comparing performance across different impact orders, since the\n magnitude of the impact will always be the same.\n\n event_series: input of shape (T,) with event occurrences\n order: order of the event impacts\n const: constant for mean shift'''\n\n series_length = len(event_series)\n time_series = np.random.randn(series_length)\n for t in range(series_length):\n if event_series[t] == 1:\n time_series[t+1:t+order+1] += const\n return time_series\n\[email protected]\ndef time_series_var_impact(event_series, order, variance):\n '''Generate a time series with impacts in variance as described in the paper.\n\n event_series: input of shape (T,) with event occurrences\n order: order of the event impacts\n variance: variance under event impacts'''\n\n series_length = len(event_series)\n time_series = np.random.randn(series_length)\n for t in range(series_length):\n if event_series[t] == 1:\n for tt in range(t+1, min(series_length, t+order+1)):\n time_series[tt] = np.random.randn()*np.sqrt(variance)\n return time_series\n\[email protected]\ndef time_series_tail_impact(event_series, order, dof):\n '''Generate a time series with impacts in tails as described in the paper.\n\n event_series: input of shape (T,) with event occurrences\n order: delay of the event impacts\n dof: degrees of freedom of the t distribution'''\n\n series_length = len(event_series)\n time_series = np.random.randn(series_length)*np.sqrt(dof/(dof-2))\n for t in range(series_length):\n if event_series[t] == 1:\n for tt in range(t+1, min(series_length, t+order+1)):\n time_series[tt] = np.random.standard_t(dof)\n return time_series",
"_____no_output_____"
]
],
[
[
"# Visualization of the impact models",
"_____no_output_____"
]
],
[
[
"default_T = 8192\ndefault_N = 64\ndefault_q = 4\n\nes = event_series_bernoulli(default_T, default_N)\n\nfor ts in [\n time_series_mean_impact(es, order=default_q, signal_to_noise=10.),\n time_series_meanconst_impact(es, order=default_q, const=5.),\n time_series_var_impact(es, order=default_q, variance=4.),\n time_series_tail_impact(es, order=default_q, dof=3.),\n]:\n fig, (ax1, ax2) = plt.subplots(1, 2, gridspec_kw={'width_ratios': [2, 1]}, figsize=(15, 2))\n ax1.plot(ts)\n ax1.plot(es*np.max(ts), alpha=0.5)\n ax1.set_xlim(0, len(es))\n samples = eitest.obtain_samples(es, ts, method='eager', lag_cutoff=15, instantaneous=True)\n eitest.plot_samples(samples, ax2)\n plt.show()",
"_____no_output_____"
]
],
[
[
"# Simulations",
"_____no_output_____"
]
],
[
[
"def test_simul_pairs(impact_model, param_T, param_N, param_q, param_r,\n n_pairs, lag_cutoff, instantaneous, sample_method,\n twosamp_test, multi_test, alpha):\n true_positive = 0.\n false_positive = 0.\n for _ in tqdm(range(n_pairs)):\n es = event_series_bernoulli(param_T, param_N)\n if impact_model == 'mean':\n ts = time_series_mean_impact(es, param_q, param_r)\n elif impact_model == 'meanconst':\n ts = time_series_meanconst_impact(es, param_q, param_r)\n elif impact_model == 'var':\n ts = time_series_var_impact(es, param_q, param_r)\n elif impact_model == 'tail':\n ts = time_series_tail_impact(es, param_q, param_r)\n else:\n raise ValueError('impact_model must be \"mean\", \"meanconst\", \"var\" or \"tail\"')\n\n # coupled pair\n samples = eitest.obtain_samples(es, ts, lag_cutoff=lag_cutoff,\n method=sample_method,\n instantaneous=instantaneous,\n sort=(twosamp_test == 'ks')) # samples need to be sorted for K-S test\n tstats, pvals = eitest.pairwise_twosample_tests(samples, twosamp_test, min_pts=2)\n pvals_adj = eitest.multitest(np.sort(pvals[~np.isnan(pvals)]), multi_test)\n true_positive += (pvals_adj.min() < alpha)\n\n # uncoupled pair\n samples = eitest.obtain_samples(np.random.permutation(es), ts, lag_cutoff=lag_cutoff,\n method=sample_method,\n instantaneous=instantaneous,\n sort=(twosamp_test == 'ks'))\n tstats, pvals = eitest.pairwise_twosample_tests(samples, twosamp_test, min_pts=2)\n pvals_adj = eitest.multitest(np.sort(pvals[~np.isnan(pvals)]), multi_test)\n false_positive += (pvals_adj.min() < alpha)\n \n return true_positive/n_pairs, false_positive/n_pairs",
"_____no_output_____"
],
[
"# global parameters\n\ndefault_T = 8192\nn_pairs = 100\nalpha = 0.05\ntwosamp_test = 'ks'\nmulti_test = 'simes'\nsample_method = 'lazy'\nlag_cutoff = 32\ninstantaneous = True",
"_____no_output_____"
]
],
[
[
"## Mean impact model",
"_____no_output_____"
]
],
[
[
"default_N = 64\ndefault_r = 1.\ndefault_q = 4",
"_____no_output_____"
]
],
[
[
"### ... by number of events",
"_____no_output_____"
]
],
[
[
"vals = [4, 8, 16, 32, 64, 128, 256]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='mean', param_T=default_T,\n param_N=val, param_q=default_q, param_r=default_r,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_N, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# mean impact model (T={default_T}, q={default_q}, r={default_r}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# N\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:05<00:00, 18.73it/s]\n100%|██████████| 100/100 [00:00<00:00, 451.99it/s]\n100%|██████████| 100/100 [00:00<00:00, 439.85it/s]\n100%|██████████| 100/100 [00:00<00:00, 379.15it/s]\n100%|██████████| 100/100 [00:00<00:00, 276.60it/s]\n100%|██████████| 100/100 [00:00<00:00, 163.88it/s]\n100%|██████████| 100/100 [00:01<00:00, 78.51it/s]\n"
]
],
[
[
"### ... by impact order",
"_____no_output_____"
]
],
[
[
"vals = [1, 2, 4, 8, 16, 32]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='mean', param_T=default_T,\n param_N=default_N, param_q=val, param_r=default_r,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_q, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# mean impact model (T={default_T}, N={default_N}, r={default_r}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# q\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:00<00:00, 218.61it/s]\n100%|██████████| 100/100 [00:00<00:00, 187.72it/s]\n100%|██████████| 100/100 [00:00<00:00, 207.15it/s]\n100%|██████████| 100/100 [00:00<00:00, 200.33it/s]\n100%|██████████| 100/100 [00:00<00:00, 213.18it/s]\n100%|██████████| 100/100 [00:00<00:00, 215.75it/s]\n"
]
],
[
[
"### ... by signal-to-noise ratio",
"_____no_output_____"
]
],
[
[
"vals = [1./32, 1./16, 1./8, 1./4, 1./2, 1., 2., 4.]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='mean', param_T=default_T,\n param_N=default_N, param_q=default_q, param_r=val,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_r, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# mean impact model (T={default_T}, N={default_N}, q={default_q}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# r\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')",
"100%|██████████| 100/100 [00:00<00:00, 179.47it/s]\n100%|██████████| 100/100 [00:00<00:00, 210.34it/s]\n100%|██████████| 100/100 [00:00<00:00, 206.91it/s]\n100%|██████████| 100/100 [00:00<00:00, 214.85it/s]\n100%|██████████| 100/100 [00:00<00:00, 212.98it/s]\n100%|██████████| 100/100 [00:00<00:00, 182.82it/s]\n100%|██████████| 100/100 [00:00<00:00, 181.18it/s]\n100%|██████████| 100/100 [00:00<00:00, 210.13it/s]\n"
]
],
[
[
"## Meanconst impact model",
"_____no_output_____"
]
],
[
[
"default_N = 64\ndefault_r = 0.5\ndefault_q = 4",
"_____no_output_____"
]
],
[
[
"### ... by number of events",
"_____no_output_____"
]
],
[
[
"vals = [4, 8, 16, 32, 64, 128, 256]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='meanconst', param_T=default_T,\n param_N=val, param_q=default_q, param_r=default_r,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_N, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# meanconst impact model (T={default_T}, q={default_q}, r={default_r}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# N\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:00<00:00, 370.92it/s]\n100%|██████████| 100/100 [00:00<00:00, 387.87it/s]\n100%|██████████| 100/100 [00:00<00:00, 364.85it/s]\n100%|██████████| 100/100 [00:00<00:00, 313.86it/s]\n100%|██████████| 100/100 [00:00<00:00, 215.43it/s]\n100%|██████████| 100/100 [00:00<00:00, 115.63it/s]\n100%|██████████| 100/100 [00:01<00:00, 52.62it/s]\n"
]
],
[
[
"### ... by impact order",
"_____no_output_____"
]
],
[
[
"vals = [1, 2, 4, 8, 16, 32]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='meanconst', param_T=default_T,\n param_N=default_N, param_q=val, param_r=default_r,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_q, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# meanconst impact model (T={default_T}, N={default_N}, r={default_r}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# q\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:00<00:00, 191.97it/s]\n100%|██████████| 100/100 [00:00<00:00, 209.09it/s]\n100%|██████████| 100/100 [00:00<00:00, 181.51it/s]\n100%|██████████| 100/100 [00:00<00:00, 170.74it/s]\n100%|██████████| 100/100 [00:00<00:00, 196.70it/s]\n100%|██████████| 100/100 [00:00<00:00, 191.42it/s]\n"
]
],
[
[
"### ... by mean value",
"_____no_output_____"
]
],
[
[
"vals = [0.125, 0.25, 0.5, 1, 2]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='meanconst', param_T=default_T,\n param_N=default_N, param_q=default_q, param_r=val,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_r, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# meanconst impact model (T={default_T}, N={default_N}, q={default_q}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# r\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:00<00:00, 172.66it/s]\n100%|██████████| 100/100 [00:00<00:00, 212.73it/s]\n100%|██████████| 100/100 [00:00<00:00, 210.24it/s]\n100%|██████████| 100/100 [00:00<00:00, 153.75it/s]\n100%|██████████| 100/100 [00:00<00:00, 211.59it/s]\n"
]
],
[
[
"## Variance impact model\nIn the paper, we show results with the variance impact model parametrized by the **variance increase**. Here we directly modulate the variance.",
"_____no_output_____"
]
],
[
[
"default_N = 64\ndefault_r = 8.\ndefault_q = 4",
"_____no_output_____"
]
],
[
[
"### ... by number of events",
"_____no_output_____"
]
],
[
[
"vals = [4, 8, 16, 32, 64, 128, 256]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='var', param_T=default_T,\n param_N=val, param_q=default_q, param_r=default_r,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_N, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# var impact model (T={default_T}, q={default_q}, r={default_r}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# N\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:00<00:00, 379.83it/s]\n100%|██████████| 100/100 [00:00<00:00, 399.36it/s]\n100%|██████████| 100/100 [00:00<00:00, 372.13it/s]\n100%|██████████| 100/100 [00:00<00:00, 319.38it/s]\n100%|██████████| 100/100 [00:00<00:00, 216.67it/s]\n100%|██████████| 100/100 [00:00<00:00, 121.62it/s]\n100%|██████████| 100/100 [00:01<00:00, 58.75it/s]\n"
]
],
[
[
"### ... by impact order",
"_____no_output_____"
]
],
[
[
"vals = [1, 2, 4, 8, 16, 32]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='var', param_T=default_T,\n param_N=default_N, param_q=val, param_r=default_r,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_q, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# var impact model (T={default_T}, N={default_N}, r={default_r}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# q\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:00<00:00, 205.11it/s]\n100%|██████████| 100/100 [00:00<00:00, 208.57it/s]\n100%|██████████| 100/100 [00:00<00:00, 208.42it/s]\n100%|██████████| 100/100 [00:00<00:00, 215.50it/s]\n100%|██████████| 100/100 [00:00<00:00, 210.17it/s]\n100%|██████████| 100/100 [00:00<00:00, 213.72it/s]\n"
]
],
[
[
"### ... by variance",
"_____no_output_____"
]
],
[
[
"vals = [2., 4., 8., 16., 32.]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='var', param_T=default_T,\n param_N=default_N, param_q=default_q, param_r=val,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_r, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# var impact model (T={default_T}, N={default_N}, q={default_q}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# r\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:00<00:00, 211.99it/s]\n100%|██████████| 100/100 [00:00<00:00, 213.48it/s]\n100%|██████████| 100/100 [00:00<00:00, 209.49it/s]\n100%|██████████| 100/100 [00:00<00:00, 214.06it/s]\n100%|██████████| 100/100 [00:00<00:00, 213.53it/s]\n"
]
],
[
[
"## Tail impact model",
"_____no_output_____"
]
],
[
[
"default_N = 512\ndefault_r = 3.\ndefault_q = 4",
"_____no_output_____"
]
],
[
[
"### ... by number of events",
"_____no_output_____"
]
],
[
[
"vals = [64, 128, 256, 512, 1024]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='tail', param_T=default_T,\n param_N=val, param_q=default_q, param_r=default_r,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_N, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# tail impact model (T={default_T}, q={default_q}, r={default_r}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# N\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:00<00:00, 210.81it/s]\n100%|██████████| 100/100 [00:00<00:00, 117.61it/s]\n100%|██████████| 100/100 [00:01<00:00, 58.35it/s]\n100%|██████████| 100/100 [00:03<00:00, 26.73it/s]\n100%|██████████| 100/100 [00:07<00:00, 13.43it/s]\n"
]
],
[
[
"### ... by impact order",
"_____no_output_____"
]
],
[
[
"vals = [1, 2, 4, 8, 16, 32]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='tail', param_T=default_T,\n param_N=default_N, param_q=val, param_r=default_r,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_q, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.gca().set_xscale('log', base=2)\nplt.legend()\nplt.show()\n\nprint(f'# tail impact model (T={default_T}, N={default_N}, r={default_r}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# q\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:03<00:00, 28.23it/s]\n100%|██████████| 100/100 [00:03<00:00, 27.89it/s]\n100%|██████████| 100/100 [00:03<00:00, 28.22it/s]\n100%|██████████| 100/100 [00:03<00:00, 27.32it/s]\n100%|██████████| 100/100 [00:03<00:00, 27.25it/s]\n100%|██████████| 100/100 [00:03<00:00, 26.63it/s]\n"
]
],
[
[
"### ... by degrees of freedom",
"_____no_output_____"
]
],
[
[
"vals = [2.5, 3., 3.5, 4., 4.5, 5., 5.5, 6.]\n\ntprs = np.empty(len(vals))\nfprs = np.empty(len(vals))\nfor i, val in enumerate(vals):\n tprs[i], fprs[i] = test_simul_pairs(impact_model='tail', param_T=default_T,\n param_N=default_N, param_q=default_q, param_r=val,\n n_pairs=n_pairs, sample_method=sample_method,\n lag_cutoff=lag_cutoff, instantaneous=instantaneous,\n twosamp_test=twosamp_test, multi_test=multi_test, alpha=alpha)\n\nplt.figure(figsize=(3,3))\nplt.axvline(default_r, ls='-', c='gray', lw=1, label='def')\nplt.axhline(alpha, ls='--', c='black', lw=1, label='alpha')\nplt.plot(vals, tprs, label='TPR', marker='x')\nplt.plot(vals, fprs, label='FPR', marker='x')\nplt.legend()\nplt.show()\n\nprint(f'# tail impact model (T={default_T}, N={default_N}, q={default_q}, n_pairs={n_pairs}, cutoff={lag_cutoff}, instantaneous={instantaneous}, alpha={alpha}, {sample_method}-{twosamp_test}-{multi_test})')\nprint(f'# r\\ttpr\\tfpr')\nfor i, (tpr, fpr) in enumerate(zip(tprs, fprs)):\n print(f'{vals[i]}\\t{tpr}\\t{fpr}')\nprint()",
"100%|██████████| 100/100 [00:03<00:00, 27.68it/s]\n100%|██████████| 100/100 [00:03<00:00, 27.97it/s]\n100%|██████████| 100/100 [00:03<00:00, 27.91it/s]\n100%|██████████| 100/100 [00:03<00:00, 28.07it/s]\n100%|██████████| 100/100 [00:03<00:00, 27.99it/s]\n100%|██████████| 100/100 [00:03<00:00, 27.71it/s]\n100%|██████████| 100/100 [00:03<00:00, 27.94it/s]\n100%|██████████| 100/100 [00:03<00:00, 27.64it/s]\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d002b497b780d100d2744ba4e601610df2c25117 | 569,608 | ipynb | Jupyter Notebook | examples/Top 25 AY Graph w Roster and Team Logo Data.ipynb | AccidentalGuru/nflfastpy | c16dcc13dc91c6a051f39ea8c28962a789322762 | [
"MIT"
] | null | null | null | examples/Top 25 AY Graph w Roster and Team Logo Data.ipynb | AccidentalGuru/nflfastpy | c16dcc13dc91c6a051f39ea8c28962a789322762 | [
"MIT"
] | null | null | null | examples/Top 25 AY Graph w Roster and Team Logo Data.ipynb | AccidentalGuru/nflfastpy | c16dcc13dc91c6a051f39ea8c28962a789322762 | [
"MIT"
] | null | null | null | 1,600.022472 | 546,150 | 0.956005 | [
[
[
"<a href=\"https://colab.research.google.com/github/AccidentalGuru/nflfastpy/blob/master/examples/Top%2025%20AY%20Graph%20w%20Roster%20and%20Team%20Logo%20Data.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"%%capture\n%pip install nflfastpy --upgrade",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"import nflfastpy\nfrom nflfastpy.utils import convert_to_gsis_id\nfrom nflfastpy import default_headshot\nfrom matplotlib import pyplot as plt\nimport pandas as pd\nimport seaborn as sns\nimport requests",
"_____no_output_____"
],
[
"print('Example default player headshot\\n')\nplt.imshow(default_headshot);",
"Example default player headshot\n\n"
],
[
"df = nflfastpy.load_pbp_data(year=2020)\nroster_df = nflfastpy.load_roster_data()\nteam_logo_df = nflfastpy.load_team_logo_data()",
"_____no_output_____"
],
[
"roster_df = roster_df.loc[roster_df['team.season'] == 2019]\n\nair_yards_df = df.loc[df['pass_attempt'] == 1, ['receiver_player_id', 'receiver_player_name', 'posteam', 'air_yards']]\n\nair_yards_df = air_yards_df.loc[air_yards_df['receiver_player_id'].notnull()]\n\nair_yards_df['gsis_id'] = air_yards_df['receiver_player_id'].apply(convert_to_gsis_id)\n\n#grabbing the top 5 air yards\ntop_25 = air_yards_df.groupby('gsis_id')['air_yards'].sum().sort_values(ascending=False)[:25].index.unique()\n\nair_yards_df = air_yards_df.loc[air_yards_df['gsis_id'].isin(top_25)]\n\nair_yards_df.head()",
"_____no_output_____"
],
[
"air_yards_df['receiver_player_name'].unique()",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(25, 2, figsize=(20, 40))\n\nfor i, row in enumerate(axes):\n \n ax1, ax2 = row[0], row[1]\n \n player_gsis_id = top_25[i]\n player_df = air_yards_df.loc[air_yards_df['gsis_id'] == player_gsis_id]\n team_logo_data = team_logo_df.loc[team_logo_df['team_abbr'] == player_df['posteam'].values[0]]\n team_color_1 = team_logo_data['team_color'].values[0]\n team_color_2 = team_logo_data['team_color2'].values[0]\n\n player_roster_data = roster_df.loc[roster_df['teamPlayers.gsisId'] == player_gsis_id]\n\n if player_roster_data.empty:\n #if the player is a rookie\n a = default_headshot\n else:\n player_headshot = player_roster_data['teamPlayers.headshot_url'].values[0]\n a = plt.imread(player_headshot)\n \n ax1.set_title(player_df['receiver_player_name'].values[0])\n ax1.imshow(a)\n ax1.axis('off')\n sns.kdeplot(player_df['air_yards'], color=team_color_2, ax=ax2)\n x = ax2.get_lines()[0].get_xydata()[:, 0]\n y = ax2.get_lines()[0].get_xydata()[:, 1]\n ax2.set_xticks(range(-10, 60, 10))\n ax2.fill_between(x, y, color=team_color_1, alpha=0.5)\n\nplt.show();",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d002bc0e0081d73349f836a6e32db713d13f5fa2 | 948,447 | ipynb | Jupyter Notebook | Analyses/The Evolution of The Russian Comedy_Verse_Features.ipynb | innawendell/European_Comedy | f9f6bf2844878503bccb9de2efe549ffc9c7df6b | [
"MIT"
] | null | null | null | Analyses/The Evolution of The Russian Comedy_Verse_Features.ipynb | innawendell/European_Comedy | f9f6bf2844878503bccb9de2efe549ffc9c7df6b | [
"MIT"
] | null | null | null | Analyses/The Evolution of The Russian Comedy_Verse_Features.ipynb | innawendell/European_Comedy | f9f6bf2844878503bccb9de2efe549ffc9c7df6b | [
"MIT"
] | null | null | null | 383.056139 | 157,244 | 0.922239 | [
[
[
"## The Analysis of The Evolution of The Russian Comedy. Part 3.",
"_____no_output_____"
],
[
"In this analysis,we will explore evolution of the French five-act comedy in verse based on the following features:\n\n- The coefficient of dialogue vivacity;\n- The percentage of scenes with split verse lines;\n- The percentage of scenes with split rhymes;\n- The percentage of open scenes.\n- The percentage of scenes with split verse lines and rhymes.\n\nWe will tackle the following questions:\n1. We will describe the features;\n2. We will explore feature correlations.\n3. We will check the features for normality using Shapiro-Wilk normality test. This will help us determine whether parametric vs. non-parametric statistical tests are more appropriate. If the features are not normally distributed, we will use non-parametric tests. \n4. In our previous analysis of Sperantov's data, we discovered that instead of four periods of the Russian five-act tragedy in verse proposed by Sperantov, we can only be confident in the existence of two periods, where 1795 is the cut-off year. Therefore, we propose the following periods for the Russian verse comedy:\n - Period One (from 1775 to 1794)\n - Period Two (from 1795 to 1849).\n5. We will run statistical tests to determine whether these two periods are statistically different.\n6. We will create visualizations for each feature.\n7. We will run descriptive statistics for each feature.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport json\nfrom os import listdir\nfrom scipy.stats import shapiro\nimport matplotlib.pyplot as plt\n%matplotlib inline \nimport seaborn as sns",
"_____no_output_____"
],
[
"def make_plot(feature, title):\n mean, std, median = summary(feature)\n plt.figure(figsize=(10, 7))\n plt.title(title, fontsize=17)\n sns.distplot(feature, kde=False)\n mean_line = plt.axvline(mean, \n color='black',\n linestyle='solid', \n linewidth=2); M1 = 'Mean';\n median_line = plt.axvline(median, \n color='green',linestyle='dashdot', \n linewidth=2); M2='Median'\n std_line = plt.axvline(mean + std, \n color='black',\n linestyle='dashed', \n linewidth=2); M3 = 'Standard deviation';\n plt.axvline(mean - std, \n color='black',\n linestyle='dashed', \n linewidth=2)\n\n plt.legend([mean_line, median_line, std_line], [M1, M2, M3])\n plt.show()",
"_____no_output_____"
],
[
"def small_sample_mann_whitney_u_test(series_one, series_two):\n values_one = series_one.sort_values().tolist()\n values_two = series_two.sort_values().tolist()\n # make sure there are no ties - this function only works for no ties\n result_df = pd.DataFrame(values_one + values_two, columns=['combined']).sort_values(by='combined')\n # average for ties\n result_df['ranks'] = result_df['combined'].rank(method='average')\n # make a dictionary where keys are values and values are ranks\n val_to_rank = dict(zip(result_df['combined'].values, result_df['ranks'].values))\n sum_ranks_one = np.sum([val_to_rank[num] for num in values_one])\n sum_ranks_two = np.sum([val_to_rank[num] for num in values_two])\n # number in sample one and two\n n_one = len(values_one) \n n_two = len(values_two)\n # calculate the mann whitney u statistic which is the smaller of the u_one and u_two\n u_one = ((n_one * n_two) + (n_one * (n_one + 1) / 2)) - sum_ranks_one\n u_two = ((n_one * n_two) + (n_two * (n_two + 1) / 2)) - sum_ranks_two \n # add a quality check \n assert u_one + u_two == n_one * n_two\n u_statistic = np.min([u_one, u_two])\n \n return u_statistic",
"_____no_output_____"
],
[
"def summary(feature):\n mean = feature.mean()\n std = feature.std()\n median = feature.median()\n \n return mean, std, median",
"_____no_output_____"
],
[
"# updated boundaries\ndef determine_period(row):\n if row <= 1794:\n period = 1\n else:\n period = 2\n \n return period",
"_____no_output_____"
]
],
[
[
"## Part 1. Feature Descriptions",
"_____no_output_____"
],
[
"For the Russian corpus of the five-act comedies, we generated additional features that inspired by Iarkho. So far, we had no understanding how these features evolved over time and whether they could differentiate literary periods.\n \nThe features include the following:\n1. **The Coefficient of Dialogue Vivacity**, i.e., the number of utterances in a play / the number of verse lines in a play. Since some of the comedies in our corpus were written in iambic hexameter while others were written in free iambs, it is important to clarify how we made sure the number of verse lines was comparable. Because Aleksandr Griboedov's *Woe From Wit* is the only four-act comedy in verse that had an extensive markup, we used it as the basis for our calculation. \n - First, improved the Dracor's markup of the verse lines in *Woe From Wit*.\n - Next, we calculated the number of verse lines in *Woe From Wit*, which was 2220.\n - Then, we calculated the total number of syllables in *Woe From Wit*, which was 22076.\n - We calculated the average number of syllables per verse line: 22076 / 2220 = 9.944144144144143.\n - Finally, we divided the average number of syllables in *Woe From Wit* by the average number of syllables in a comedy written in hexameter, i.e., 12.5: 9.944144144144143 / 12.5 = 0.796.\n - To convert the number of verse lines in a play written in free iambs and make it comparable with the comedies written in hexameter, we used the following formula: rescaled_number of verse lines = the number of verse lines in free iambs * 0.796. \n - For example, in *Woe From Wit*, the number of verse lines = 2220, the rescaled number of verse lines = 2220 * 0.796 = 1767.12. The coefficient of dialogue vivacity = 702 / 1767.12 = 0.397. \n2. **The Percentage of Scenes with Split Verse Lines**, i.e, the percentage of scenes where the end of a scene does not correspond with the end of a verse lines and the verse line extends into the next scene, e.g., \"Не бойся. Онъ блажитъ. ЯВЛЕНІЕ 3. Какъ радъ что вижу васъ.\"\n3. **The Percentage of Scenes with Split Rhymes**, i.e., the percentage of scenes that rhyme with other scenes, e.g., \"Надѣюсъ на тебя, Вѣтрана, какъ на стѣну. ЯВЛЕНІЕ 4. И въ ней , какъ ни крѣпка, мы видимЪ перемѣну.\"\n4. **The Percentage of Open Scenes**, i.e., the percentage of scenes with either split verse lines or rhymes.\n5. **The Percentage of Scenes With Split Verse Lines and Rhymes**, i.e., the percentage of scenes that are connected through both means: by sharing a verse lines and a rhyme.",
"_____no_output_____"
]
],
[
[
"comedies = pd.read_csv('../Russian_Comedies/Data/Comedies_Raw_Data.csv')",
"_____no_output_____"
],
[
"# sort by creation date\ncomedies_sorted = comedies.sort_values(by='creation_date').copy()",
"_____no_output_____"
],
[
"# select only original comedies and five act\noriginal_comedies = comedies_sorted[(comedies_sorted['translation/adaptation'] == 0) & \n (comedies_sorted['num_acts'] == 5)].copy()",
"_____no_output_____"
],
[
"original_comedies.head()",
"_____no_output_____"
],
[
"original_comedies.shape",
"_____no_output_____"
],
[
"# rename column names for clarity\noriginal_comedies = original_comedies.rename(columns={'num_scenes_iarkho': 'mobility_coefficient'})",
"_____no_output_____"
],
[
"comedies_verse_features = original_comedies[['index',\n 'title',\n 'first_name',\n 'last_name',\n 'creation_date',\n 'dialogue_vivacity',\n 'percentage_scene_split_verse',\n 'percentage_scene_split_rhymes',\n 'percentage_open_scenes',\n 'percentage_scenes_rhymes_split_verse']].copy()",
"_____no_output_____"
],
[
"comedies_verse_features.head()",
"_____no_output_____"
]
],
[
[
"## Part 1. Feature Correlations",
"_____no_output_____"
]
],
[
[
"comedies_verse_features[['dialogue_vivacity',\n 'percentage_scene_split_verse',\n 'percentage_scene_split_rhymes',\n 'percentage_open_scenes',\n 'percentage_scenes_rhymes_split_verse']].corr().round(2)",
"_____no_output_____"
],
[
"original_comedies[['dialogue_vivacity',\n 'mobility_coefficient']].corr()",
"_____no_output_____"
]
],
[
[
"Dialogue vivacity is moderately positively correlated with the percentage of scenes with split verse lines (0.53), with the percentage of scenes with split rhymes (0.51), and slightly less correlated with the percentage of open scenes (0.45). However, it is strongly positively correlated with the percentage of scenes with both split rhymes and verse lines (0.73). The scenes with very fast-paced dialogue are more likely to be interconnected through both rhyme and shared verse lines. One unexpected discovery is that dialogue vivacity only weakly correlated with the mobility coefficient (0.06): more active movement of dramatic characters on stage does not necessarily entail that their utterances are going to be shorter.\n\nThe percentage of scenes with split verse lines is moderately positively correlated with the percentage of scenes with split rhymes (0.66): the scenes that are connected by verse are likely but not necessarily always going to be connected through rhyme.\n\nSuch features as the percentage of open scenes and the percentage of scenes with split rhymes and verse lines are strongly positively correlated with their constituent features (the correlation of the percentage of open scenes with the percentage of scenes with split verse lines is 0.86, with the percentage of split rhymes is 0.92). From this, we can infer that the bulk of the open scenes are connected through rhymes. The percentage of scenes with split rhymes and verse lines is strongly positively correlated with the percentage of scenes with split verse lines (0.87) and the percentage of scenes with split rhymes.",
"_____no_output_____"
],
[
"## Part 3. Feature Distributions and Normality",
"_____no_output_____"
]
],
[
[
"make_plot(comedies_verse_features['dialogue_vivacity'], \n 'Distribution of the Dialogue Vivacity Coefficient')",
"/opt/anaconda3/envs/text_extraction/lib/python3.7/site-packages/seaborn/distributions.py:2551: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n"
],
[
"mean, std, median = summary(comedies_verse_features['dialogue_vivacity'])\nprint('Mean dialogue vivacity coefficient', round(mean, 2))\nprint('Standard deviation of the dialogue vivacity coefficient:', round(std, 2))\nprint('Median dialogue vivacity coefficient:', median)",
"Mean dialogue vivacity coefficient 0.46\nStandard deviation of the dialogue vivacity coefficient: 0.1\nMedian dialogue vivacity coefficient: 0.4575\n"
]
],
[
[
"### Shapiro-Wilk Normality Test",
"_____no_output_____"
]
],
[
[
"print('The p-value of the Shapiro-Wilk normality test:', \n shapiro(comedies_verse_features['dialogue_vivacity'])[1])",
"The p-value of the Shapiro-Wilk normality test: 0.2067030817270279\n"
]
],
[
[
"The Shapiro-Wilk test showed that the probability of the coefficient of dialogue vivacity of being normally distributed was 0.2067030817270279, which was above the 0.05 significance level. We failed to reject the null hypothesis of the normal distribution.",
"_____no_output_____"
]
],
[
[
"make_plot(comedies_verse_features['percentage_scene_split_verse'], \n 'Distribution of The Percentage of Scenes with Split Verse Lines')",
"_____no_output_____"
],
[
"mean, std, median = summary(comedies_verse_features['percentage_scene_split_verse'])\nprint('Mean percentage of scenes with split verse lines:', round(mean, 2))\nprint('Standard deviation of the percentage of scenes with split verse lines:', round(std, 2))\nprint('Median percentage of scenes with split verse lines:', median)",
"Mean percentage of scenes with split verse lines: 30.39\nStandard deviation of the percentage of scenes with split verse lines: 14.39\nMedian percentage of scenes with split verse lines: 28.854\n"
],
[
"print('The p-value of the Shapiro-Wilk normality test:', \n shapiro(comedies_verse_features['percentage_scene_split_verse'])[1])",
"The p-value of the Shapiro-Wilk normality test: 0.8681985139846802\n"
]
],
[
[
"The Shapiro-Wilk showed that the probability of the percentage of scenes with split verse lines of being normally distributed was very high (the p-value is 0.8681985139846802). We failed to reject the null hypothesis of normal distribution.",
"_____no_output_____"
]
],
[
[
"make_plot(comedies_verse_features['percentage_scene_split_rhymes'], \n 'Distribution of The Percentage of Scenes with Split Rhymes')",
"_____no_output_____"
],
[
"mean, std, median = summary(comedies_verse_features['percentage_scene_split_rhymes'])\nprint('Mean percentage of scenes with split rhymes:', round(mean, 2))\nprint('Standard deviation of the percentage of scenes with split rhymes:', round(std, 2))\nprint('Median percentage of scenes with split rhymes:', median)",
"Mean percentage of scenes with split rhymes: 39.77\nStandard deviation of the percentage of scenes with split rhymes: 16.24\nMedian percentage of scenes with split rhymes: 36.6365\n"
],
[
"print('The p-value of the Shapiro-Wilk normality test:', \n shapiro(comedies_verse_features['percentage_scene_split_rhymes'])[1])",
"The p-value of the Shapiro-Wilk normality test: 0.5752763152122498\n"
]
],
[
[
"The Shapiro-Wilk test showed that the probability of the number of dramatic characters of being normally distributed was 0.5752763152122498. This probability was much higher than the 0.05 significance level. Therefore, we failed to reject the null hypothesis of normal distribution.",
"_____no_output_____"
]
],
[
[
"make_plot(comedies_verse_features['percentage_open_scenes'], \n 'Distribution of The Percentage of Open Scenes')",
"_____no_output_____"
],
[
"mean, std, median = summary(comedies_verse_features['percentage_open_scenes'])\nprint('Mean percentage of open scenes:', round(mean, 2))\nprint('Standard deviation of the percentage of open scenes:', round(std, 2))\nprint('Median percentage of open scenes:', median)",
"Mean percentage of open scenes: 55.62\nStandard deviation of the percentage of open scenes: 19.25\nMedian percentage of open scenes: 56.6605\n"
],
[
"print('The p-value of the Shapiro-Wilk normality test:', \n shapiro(comedies_verse_features['percentage_open_scenes'])[1])",
"The p-value of the Shapiro-Wilk normality test: 0.3018988370895386\n"
]
],
[
[
"The Shapiro-Wilk test showed that the probability of the number of the percentage of open scenes of being normally distributed was 0.3018988370895386, which was quite a lot higher than the significance level of 0.05. Therefore, we failed to reject the null hypothesis of normal distribution of the percentage of open scenes.",
"_____no_output_____"
]
],
[
[
"make_plot(comedies_verse_features['percentage_scenes_rhymes_split_verse'], \n 'Distribution of The Percentage of Scenes with Split Verse Lines and Rhymes')",
"_____no_output_____"
],
[
"mean, std, median = summary(comedies_verse_features['percentage_scenes_rhymes_split_verse'])\nprint('Mean percentage of scenes with split rhymes and verse lines:', round(mean, 2))\nprint('Standard deviation of the percentage of scenes with split rhymes and verse lines:', round(std, 2))\nprint('Median percentage of scenes with split rhymes and verse lines:', median)",
"Mean percentage of scenes with split rhymes and verse lines: 14.53\nStandard deviation of the percentage of scenes with split rhymes and verse lines: 9.83\nMedian percentage of scenes with split rhymes and verse lines: 13.0155\n"
],
[
"print('The p-value of the Shapiro-Wilk normality test:', \n shapiro(comedies_verse_features['percentage_scenes_rhymes_split_verse'])[1])",
"The p-value of the Shapiro-Wilk normality test: 0.015218793414533138\n"
]
],
[
[
"The Shapiro-Wilk test showed that the probability of the percentage of scenes with split verse lines and rhymes of being normally distributed was very low (the p-value was 0.015218793414533138). Therefore, we rejected the hypothesis of normal distribution.",
"_____no_output_____"
],
[
"### Summary:\n1. The majority of the verse features were normally distributed. For them, we could use a parametric statistical test.\n2. The only feature that was not normally distributed was the percentage of scenes with split rhymes and verse lines. For this feature, we used a non-parametric test such as the Mann-Whitney u test.",
"_____no_output_____"
],
[
"## Part 3. Hypothesis Testing",
"_____no_output_____"
],
[
"We will run statistical tests to determine whether the two periods distinguishable for the Russian five-act verse tragedy are significantly different for the Russian five-act comedy. The two periods are:\n - Period One (from 1747 to 1794)\n - Period Two (from 1795 to 1822)\n \nFor all features that were normally distributed, we will use *scipy.stats* Python library to run a **t-test** to check whether there is a difference between Period One and Period Two. The null hypothesis is that there is no difference between the two periods. The alternative hypothesis is that the two periods are different. Our significance level will be set at 0.05. If the p-value produced by the t-test will be below 0.05, we will reject the null hypothesis of no difference. \n\n\nFor the percentage of scenes with split rhymes and verse lines, we will run **the Mann-Whitney u-test** to check whether there is a difference between Period One and Period Two. The null hypothesis will be no difference between these periods, whereas the alternative hypothesis will be that the periods will be different.\n\nSince both periods have fewer than 20 tragedies, we cannot use the scipy's Man-Whitney u-test that requires each sample size to be at least 20 because it uses normal approximation. Instead, we will have to run Mann-Whitney U-test without a normal approximation for which we wrote a custom function. The details about the test can be found in the following resource: https://sphweb.bumc.bu.edu/otlt/mph-modules/bs/bs704_nonparametric/bs704_nonparametric4.html.\n\nOne limitation that we need to mention is the sample size. The first period has only six comedies and the second period has only ten. However, it is impossible to increase the sample size - we cannot ask the Russian playwrights of the eighteenth and nineteenth century to produce more five-act verse comedies. If there are other Russian five-act comedies of these periods, they are either unknown or not available to us.",
"_____no_output_____"
]
],
[
[
"comedies_verse_features['period'] = comedies_verse_features.creation_date.apply(determine_period)",
"_____no_output_____"
],
[
"period_one = comedies_verse_features[comedies_verse_features['period'] == 1].copy()\nperiod_two = comedies_verse_features[comedies_verse_features['period'] == 2].copy()",
"_____no_output_____"
],
[
"period_one.shape",
"_____no_output_____"
],
[
"period_two.shape",
"_____no_output_____"
]
],
[
[
"## The T-Test",
"_____no_output_____"
],
[
"### The Coefficient of Dialogue Vivacity",
"_____no_output_____"
]
],
[
[
"from scipy.stats import ttest_ind",
"_____no_output_____"
],
[
"ttest_ind(period_one['dialogue_vivacity'],\n period_two['dialogue_vivacity'], equal_var=False)",
"_____no_output_____"
]
],
[
[
"### The Percentage of Scenes With Split Verse Lines",
"_____no_output_____"
]
],
[
[
"ttest_ind(period_one['percentage_scene_split_verse'],\n period_two['percentage_scene_split_verse'], equal_var=False)",
"_____no_output_____"
]
],
[
[
"### The Percentage of Scnes With Split Rhymes",
"_____no_output_____"
]
],
[
[
"ttest_ind(period_one['percentage_scene_split_rhymes'],\n period_two['percentage_scene_split_rhymes'], equal_var=False)",
"_____no_output_____"
]
],
[
[
"### The Percentage of Open Scenes",
"_____no_output_____"
]
],
[
[
"ttest_ind(period_one['percentage_open_scenes'],\n period_two['percentage_open_scenes'], equal_var=False)",
"_____no_output_____"
]
],
[
[
"### Summary\n\n|Feature |p-value |Result\n|---------------------------| ----------------|--------------------------------\n| The coefficient of dialogue vivacity |0.92 | Not Significant\n|The percentage of scenes with split verse lines|0.009 | Significant\n|The percentage of scenes with split rhymes| 0.44| Not significant\n|The percentage of open scenes| 0.10| Not significant",
"_____no_output_____"
],
[
"## The Mann-Whitney Test",
"_____no_output_____"
],
[
"The Process:\n- Our null hypothesis is that there is no difference between two periods. Our alternative hypothesis is that the periods are different.\n- We will set the signficance level (alpha) at 0.05.\n- We will run the test and calculate the test statistic.\n- We will compare the test statistic with the critical value of U for a two-tailed test at alpha=0.05. Critical values can be found at https://www.real-statistics.com/statistics-tables/mann-whitney-table/.\n- If our test statistic is equal or lower than the critical value of U, we will reject the null hypothesis. Otherwise, we will fail to reject it.",
"_____no_output_____"
],
[
"### The Percentage of Scenes With Split Verse Lines and Rhymes",
"_____no_output_____"
]
],
[
[
"small_sample_mann_whitney_u_test(period_one['percentage_scenes_rhymes_split_verse'],\n period_two['percentage_scenes_rhymes_split_verse'])",
"_____no_output_____"
]
],
[
[
"### Critical Value of U",
"_____no_output_____"
],
[
"|Periods |Critical Value of U \n|---------------------------| ----------------\n| Period One (n=6) and Period Two (n=10) |11\n",
"_____no_output_____"
],
[
"### Summary\n\n|Feature |u-statistic |Result\n|---------------------------| ----------------|--------------------------------\n| The percentage of scenes with split verse lines and rhymes|21 | Not Significant",
"_____no_output_____"
],
[
"We discovered that the distribution of only one feature, the percentage of scenes with split verse lines, was different in Periods One and Two. Distributions of other features did not prove to be significantly different. ",
"_____no_output_____"
],
[
"## Part 4. Visualizations",
"_____no_output_____"
]
],
[
[
"def scatter(df, feature, title, xlabel, text_y):\n sns.jointplot('creation_date', \n feature,\n data=df, \n color='b', \n height=7).plot_joint(\n sns.kdeplot, \n zorder=0, \n n_levels=20)\n plt.axvline(1795, color='grey',linestyle='dashed', linewidth=2)\n plt.text(1795.5, text_y, '1795')\n plt.title(title, fontsize=20, pad=100)\n plt.xlabel('Date', fontsize=14)\n plt.ylabel(xlabel, fontsize=14)\n\n plt.show()",
"_____no_output_____"
]
],
[
[
"### The Coefficient of Dialogue Vivacity",
"_____no_output_____"
]
],
[
[
"scatter(comedies_verse_features, \n 'dialogue_vivacity', \n 'The Coefficient of Dialogue Vivacity by Year', \n 'The Coefficient of Dialogue Vivacity',\n 0.85)",
"/opt/anaconda3/envs/text_extraction/lib/python3.7/site-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
]
],
[
[
"### The Percentage of Scenes With Split Verse Lines",
"_____no_output_____"
]
],
[
[
"scatter(comedies_verse_features, \n 'percentage_scene_split_verse', \n 'The Percentage of Scenes With Split Verse Lines by Year', \n 'Percentage of Scenes With Split Verse Lines',\n 80)",
"/opt/anaconda3/envs/text_extraction/lib/python3.7/site-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
]
],
[
[
"### The Percentage of Scenes With Split Rhymes",
"_____no_output_____"
]
],
[
[
"scatter(comedies_verse_features, \n 'percentage_scene_split_rhymes', \n 'The Percentage of Scenes With Split Rhymes by Year', \n 'The Percentage of Scenes With Split Rhymes',\n 80)",
"/opt/anaconda3/envs/text_extraction/lib/python3.7/site-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
]
],
[
[
"### The Percentage of Open Scenes",
"_____no_output_____"
]
],
[
[
"scatter(comedies_verse_features, \n 'percentage_open_scenes', \n 'The Percentage of Open Scenes by Year', \n 'The Percentage of Open Scenes',\n 100)",
"/opt/anaconda3/envs/text_extraction/lib/python3.7/site-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
]
],
[
[
"### The Percentage of Scenes With Split Verse Lines and Rhymes",
"_____no_output_____"
]
],
[
[
"scatter(comedies_verse_features, \n 'percentage_scenes_rhymes_split_verse', \n ' The Percentage of Scenes With Split Verse Lines and Rhymes by Year', \n ' The Percentage of Scenes With Split Verse Lines and Rhymes',\n 45)",
"/opt/anaconda3/envs/text_extraction/lib/python3.7/site-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
]
],
[
[
"## Part 5. Descriptive Statistics For Two Periods and Overall",
"_____no_output_____"
],
[
"### The Coefficient of Dialogue Vivacity",
"_____no_output_____"
],
[
"#### In Entire Corpus",
"_____no_output_____"
]
],
[
[
"comedies_verse_features.describe().loc[:, 'dialogue_vivacity'][['mean', \n 'std', \n '50%',\n 'min', \n 'max']].round(2)",
"_____no_output_____"
]
],
[
[
"#### By Tentative Periods",
"_____no_output_____"
]
],
[
[
"comedies_verse_features.groupby('period').describe().loc[:, 'dialogue_vivacity'][['mean', \n 'std', \n '50%',\n 'min', \n 'max']].round(2)",
"_____no_output_____"
]
],
[
[
"### The Percentage of Scenes With Split Verse Lines",
"_____no_output_____"
],
[
"#### In Entire Corpus",
"_____no_output_____"
]
],
[
[
"comedies_verse_features.describe().loc[:, 'percentage_scene_split_verse'][['mean', \n 'std', \n '50%',\n 'min', \n 'max']].round(2)",
"_____no_output_____"
]
],
[
[
"#### By Periods",
"_____no_output_____"
]
],
[
[
"comedies_verse_features.groupby('period').describe().loc[:, 'percentage_scene_split_verse'][['mean', \n 'std', \n '50%',\n 'min', \n 'max']].round(2)",
"_____no_output_____"
]
],
[
[
"### The Percentage of Scenes With Split Rhymes",
"_____no_output_____"
]
],
[
[
"comedies_verse_features.describe().loc[:, 'percentage_scene_split_rhymes'][['mean', \n 'std', \n '50%',\n 'min', \n 'max']].round(2)",
"_____no_output_____"
]
],
[
[
"#### By Tentative Periods",
"_____no_output_____"
]
],
[
[
"comedies_verse_features.groupby('period').describe().loc[:, 'percentage_scene_split_rhymes'][['mean', \n 'std', \n '50%',\n 'min', \n 'max']].round(2)",
"_____no_output_____"
]
],
[
[
"### The Percentage of Open Scenes",
"_____no_output_____"
],
[
"#### In Entire Corpus",
"_____no_output_____"
]
],
[
[
"comedies_verse_features.describe().loc[:, 'percentage_open_scenes'][['mean', \n 'std', \n '50%',\n 'min', \n 'max']].round(2)",
"_____no_output_____"
]
],
[
[
"#### By Tenative Periods",
"_____no_output_____"
]
],
[
[
"comedies_verse_features.groupby('period').describe().loc[:, 'percentage_open_scenes'][['mean', \n 'std', \n '50%',\n 'min', \n 'max']].round(2)",
"_____no_output_____"
]
],
[
[
"### The Percentage of Scenes With Split Verse Lines or Rhymes",
"_____no_output_____"
]
],
[
[
"comedies_verse_features.describe().loc[:, 'percentage_scenes_rhymes_split_verse'][['mean', \n 'std', \n '50%',\n 'min', \n 'max']].round(2)",
"_____no_output_____"
],
[
"comedies_verse_features.groupby('period').describe().loc[:, 'percentage_scenes_rhymes_split_verse'][['mean', \n 'std', \n '50%',\n 'min', \n 'max']].round(2)",
"_____no_output_____"
]
],
[
[
"### Summary:\n1. The mean dialogue vivacity in the corpus of the Russian five-act comedy in verse was 0.46, with a 0.10 standard deviation. In the tentative Period One, the mean dialogue vivacity was 0.46, the same as in the tentative Period Two. The standard deviation increased from 0.05 in the tentative Period One to 0.13 in the tentative Period Two.\n2. The mean percentage of scenes with split verse lines in the corpus was 30.39%, with a standard deviation of 14.39. In Period One, the mean percentage of scenes with split verse lines was 19.37%, with a standard deviation of 10.16. In Period Two, the mean percentage of scenes with split verse lines almost doubled to 37%, with a standard deviation of 12.57%.\n3. The average percentage of scenes with split rhymes was higher in the entire corpus of the Russian five-act comedies in verse than the average percentage of scenes with split verse lines (39.77% vs. 30.39%), as was the standard deviation (16.24% vs. 14.39%). The percentage of scenes with split rhymes grew from the tentative Period One to the tentative Period Two from 35.55% to 42.30%; the standard deviation slightly increased from 15.73% to 16.82%.\n4. In the corpus, the average percentage of open scenes was 55.62%, i.e., more than half of all scenes were connected either through rhyme or verse lines. The standard deviation was 19.25%. In the tentative Period One, the percentage of open scenes was 44.65%, with a standard deviation of 19.76%. In the tentative Period Two, the percentage of open scenes increased to 62.21%, with a standard deviation of 16.50%, i.e., the standard deviation was lower in Period Two.\n5. For the corpus, only 14.53% of all scenes were connected through both rhymes and verse lines. The standard deviation of the percentage of scenes with split verse lines and rhymes was 9.83%. In the tentative Period One, the mean percentage of scenes with split verse lines and rhymes was 10.27%, with a standard deviation of 5.22%. In the tentative Period Two, the mean percentage of scenes with split verse lines and rhymes was 17.09%, with a much higher standard deviation of 11.25%.",
"_____no_output_____"
],
[
"## Conclusions:\n1. The majority of the examined features were normally distributed, except for the percentage of scenes with split verse lines and rhymes.\n2. The distribution of the percentage of scenes with split verse lines differed significantly between Period One (from 1775 to 1794) and Period Two (from 1795 to 1849)). \n2. For other verse features, there was no evidence to suggest that the two periods of the Russian five-act comedy in verse are significantly different.\n3. The mean values of all examined features (except for the vivacity coefficient) increased from tentative Period One to Period Two. The mean vivacity coefficient remained the same from the tentative Period One to Period Two. The standard deviation of all examined features (except for the percentage of open scenes) increased from Period One to Period Two.\n4. Judging by the natural clustering in the data evident from visualizations, 1805 may be a more appropriate boundary between the two time periods for comedy.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
d002c5802863aebf588359e2d82fe7676ce02717 | 54,374 | ipynb | Jupyter Notebook | Utils/dowhy/docs/source/example_notebooks/lalonde_pandas_api.ipynb | maliha93/Fairness-Analysis-Code | acf13c6e7993704fc627249fe4ada44d8b616264 | [
"MIT"
] | 2,904 | 2019-05-07T08:09:33.000Z | 2022-03-31T18:28:41.000Z | Utils/dowhy/docs/source/example_notebooks/lalonde_pandas_api.ipynb | maliha93/Fairness-Analysis-Code | acf13c6e7993704fc627249fe4ada44d8b616264 | [
"MIT"
] | 238 | 2019-05-11T02:57:22.000Z | 2022-03-31T23:47:18.000Z | Utils/dowhy/docs/source/example_notebooks/lalonde_pandas_api.ipynb | maliha93/Fairness-Analysis-Code | acf13c6e7993704fc627249fe4ada44d8b616264 | [
"MIT"
] | 527 | 2019-05-08T16:23:45.000Z | 2022-03-30T21:02:41.000Z | 54.103483 | 7,392 | 0.660775 | [
[
[
"# Lalonde Pandas API Example\nby Adam Kelleher",
"_____no_output_____"
],
[
"We'll run through a quick example using the high-level Python API for the DoSampler. The DoSampler is different from most classic causal effect estimators. Instead of estimating statistics under interventions, it aims to provide the generality of Pearlian causal inference. In that context, the joint distribution of the variables under an intervention is the quantity of interest. It's hard to represent a joint distribution nonparametrically, so instead we provide a sample from that distribution, which we call a \"do\" sample.\n\nHere, when you specify an outcome, that is the variable you're sampling under an intervention. We still have to do the usual process of making sure the quantity (the conditional interventional distribution of the outcome) is identifiable. We leverage the familiar components of the rest of the package to do that \"under the hood\". You'll notice some similarity in the kwargs for the DoSampler.\n\n## Getting the Data\n\nFirst, download the data from the LaLonde example.",
"_____no_output_____"
]
],
[
[
"import os, sys\nsys.path.append(os.path.abspath(\"../../../\"))",
"_____no_output_____"
],
[
"from rpy2.robjects import r as R\n\n%load_ext rpy2.ipython\n#%R install.packages(\"Matching\")\n%R library(Matching)\n%R data(lalonde)\n%R -o lalonde\nlalonde.to_csv(\"lalonde.csv\",index=False)",
"R[write to console]: Loading required package: MASS\n\nR[write to console]: ## \n## Matching (Version 4.9-7, Build Date: 2020-02-05)\n## See http://sekhon.berkeley.edu/matching for additional documentation.\n## Please cite software as:\n## Jasjeet S. Sekhon. 2011. ``Multivariate and Propensity Score Matching\n## Software with Automated Balance Optimization: The Matching package for R.''\n## Journal of Statistical Software, 42(7): 1-52. \n##\n\n\n"
],
[
"# the data already loaded in the previous cell. we include the import\n# here you so you don't have to keep re-downloading it.\n\nimport pandas as pd\n\nlalonde=pd.read_csv(\"lalonde.csv\")",
"_____no_output_____"
]
],
[
[
"## The `causal` Namespace",
"_____no_output_____"
],
[
"We've created a \"namespace\" for `pandas.DataFrame`s containing causal inference methods. You can access it here with `lalonde.causal`, where `lalonde` is our `pandas.DataFrame`, and `causal` contains all our new methods! These methods are magically loaded into your existing (and future) dataframes when you `import dowhy.api`.",
"_____no_output_____"
]
],
[
[
"import dowhy.api",
"_____no_output_____"
]
],
[
[
"Now that we have the `causal` namespace, lets give it a try! \n\n## The `do` Operation\n\nThe key feature here is the `do` method, which produces a new dataframe replacing the treatment variable with values specified, and the outcome with a sample from the interventional distribution of the outcome. If you don't specify a value for the treatment, it leaves the treatment untouched:",
"_____no_output_____"
]
],
[
[
"do_df = lalonde.causal.do(x='treat',\n outcome='re78',\n common_causes=['nodegr', 'black', 'hisp', 'age', 'educ', 'married'],\n variable_types={'age': 'c', 'educ':'c', 'black': 'd', 'hisp': 'd', \n 'married': 'd', 'nodegr': 'd','re78': 'c', 'treat': 'b'},\n proceed_when_unidentifiable=True)",
"WARNING:dowhy.causal_model:Causal Graph not provided. DoWhy will construct a graph based on data inputs.\nINFO:dowhy.causal_graph:If this is observed data (not from a randomized experiment), there might always be missing confounders. Adding a node named \"Unobserved Confounders\" to reflect this.\nINFO:dowhy.causal_model:Model to find the causal effect of treatment ['treat'] on outcome ['re78']\nWARNING:dowhy.causal_identifier:If this is observed data (not from a randomized experiment), there might always be missing confounders. Causal effect cannot be identified perfectly.\nINFO:dowhy.causal_identifier:Continuing by ignoring these unobserved confounders because proceed_when_unidentifiable flag is True.\nINFO:dowhy.causal_identifier:Instrumental variables for treatment and outcome:[]\nINFO:dowhy.causal_identifier:Frontdoor variables for treatment and outcome:[]\nINFO:dowhy.do_sampler:Using WeightingSampler for do sampling.\nINFO:dowhy.do_sampler:Caution: do samplers assume iid data.\n"
]
],
[
[
"Notice you get the usual output and prompts about identifiability. This is all `dowhy` under the hood!\n\nWe now have an interventional sample in `do_df`. It looks very similar to the original dataframe. Compare them:",
"_____no_output_____"
]
],
[
[
"lalonde.head()",
"_____no_output_____"
],
[
"do_df.head()",
"_____no_output_____"
]
],
[
[
"## Treatment Effect Estimation\n\nWe could get a naive estimate before for a treatment effect by doing",
"_____no_output_____"
]
],
[
[
"(lalonde[lalonde['treat'] == 1].mean() - lalonde[lalonde['treat'] == 0].mean())['re78']",
"_____no_output_____"
]
],
[
[
"We can do the same with our new sample from the interventional distribution to get a causal effect estimate",
"_____no_output_____"
]
],
[
[
"(do_df[do_df['treat'] == 1].mean() - do_df[do_df['treat'] == 0].mean())['re78']",
"_____no_output_____"
]
],
[
[
"We could get some rough error bars on the outcome using the normal approximation for a 95% confidence interval, like\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\n1.96*np.sqrt((do_df[do_df['treat'] == 1].var()/len(do_df[do_df['treat'] == 1])) + \n (do_df[do_df['treat'] == 0].var()/len(do_df[do_df['treat'] == 0])))['re78']",
"_____no_output_____"
]
],
[
[
"but note that these DO NOT contain propensity score estimation error. For that, a bootstrapping procedure might be more appropriate.",
"_____no_output_____"
],
[
"This is just one statistic we can compute from the interventional distribution of `'re78'`. We can get all of the interventional moments as well, including functions of `'re78'`. We can leverage the full power of pandas, like",
"_____no_output_____"
]
],
[
[
"do_df['re78'].describe()",
"_____no_output_____"
],
[
"lalonde['re78'].describe()",
"_____no_output_____"
]
],
[
[
"and even plot aggregations, like",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import seaborn as sns\n\nsns.barplot(data=lalonde, x='treat', y='re78')",
"_____no_output_____"
],
[
"sns.barplot(data=do_df, x='treat', y='re78')",
"_____no_output_____"
]
],
[
[
"## Specifying Interventions\n\nYou can find the distribution of the outcome under an intervention to set the value of the treatment. ",
"_____no_output_____"
]
],
[
[
"do_df = lalonde.causal.do(x={'treat': 1},\n outcome='re78',\n common_causes=['nodegr', 'black', 'hisp', 'age', 'educ', 'married'],\n variable_types={'age': 'c', 'educ':'c', 'black': 'd', 'hisp': 'd', \n 'married': 'd', 'nodegr': 'd','re78': 'c', 'treat': 'b'},\n proceed_when_unidentifiable=True)",
"WARNING:dowhy.causal_model:Causal Graph not provided. DoWhy will construct a graph based on data inputs.\nINFO:dowhy.causal_graph:If this is observed data (not from a randomized experiment), there might always be missing confounders. Adding a node named \"Unobserved Confounders\" to reflect this.\nINFO:dowhy.causal_model:Model to find the causal effect of treatment ['treat'] on outcome ['re78']\nWARNING:dowhy.causal_identifier:If this is observed data (not from a randomized experiment), there might always be missing confounders. Causal effect cannot be identified perfectly.\nINFO:dowhy.causal_identifier:Continuing by ignoring these unobserved confounders because proceed_when_unidentifiable flag is True.\nINFO:dowhy.causal_identifier:Instrumental variables for treatment and outcome:[]\nINFO:dowhy.causal_identifier:Frontdoor variables for treatment and outcome:[]\nINFO:dowhy.do_sampler:Using WeightingSampler for do sampling.\nINFO:dowhy.do_sampler:Caution: do samplers assume iid data.\n"
],
[
"do_df.head()",
"_____no_output_____"
]
],
[
[
"This new dataframe gives the distribution of `'re78'` when `'treat'` is set to `1`.",
"_____no_output_____"
],
[
"For much more detail on how the `do` method works, check the docstring:",
"_____no_output_____"
]
],
[
[
"help(lalonde.causal.do)",
"Help on method do in module dowhy.api.causal_data_frame:\n\ndo(x, method='weighting', num_cores=1, variable_types={}, outcome=None, params=None, dot_graph=None, common_causes=None, estimand_type='nonparametric-ate', proceed_when_unidentifiable=False, stateful=False) method of dowhy.api.causal_data_frame.CausalAccessor instance\n The do-operation implemented with sampling. This will return a pandas.DataFrame with the outcome\n variable(s) replaced with samples from P(Y|do(X=x)).\n \n If the value of `x` is left unspecified (e.g. as a string or list), then the original values of `x` are left in\n the DataFrame, and Y is sampled from its respective P(Y|do(x)). If the value of `x` is specified (passed with a\n `dict`, where variable names are keys, and values are specified) then the new `DataFrame` will contain the\n specified values of `x`.\n \n For some methods, the `variable_types` field must be specified. It should be a `dict`, where the keys are\n variable names, and values are 'o' for ordered discrete, 'u' for un-ordered discrete, 'd' for discrete, or 'c'\n for continuous.\n \n Inference requires a set of control variables. These can be provided explicitly using `common_causes`, which\n contains a list of variable names to control for. These can be provided implicitly by specifying a causal graph\n with `dot_graph`, from which they will be chosen using the default identification method.\n \n When the set of control variables can't be identified with the provided assumptions, a prompt will raise to the\n user asking whether to proceed. To automatically over-ride the prompt, you can set the flag\n `proceed_when_unidentifiable` to `True`.\n \n Some methods build components during inference which are expensive. To retain those components for later\n inference (e.g. successive calls to `do` with different values of `x`), you can set the `stateful` flag to `True`.\n Be cautious about using the `do` operation statefully. State is set on the namespace, rather than the method, so\n can behave unpredictably. To reset the namespace and run statelessly again, you can call the `reset` method.\n \n :param x: str, list, dict: The causal state on which to intervene, and (optional) its interventional value(s).\n :param method: The inference method to use with the sampler. Currently, `'mcmc'`, `'weighting'`, and\n `'kernel_density'` are supported. The `mcmc` sampler requires `pymc3>=3.7`.\n :param num_cores: int: if the inference method only supports sampling a point at a time, this will parallelize\n sampling.\n :param variable_types: dict: The dictionary containing the variable types. Must contain the union of the causal\n state, control variables, and the outcome.\n :param outcome: str: The outcome variable.\n :param params: dict: extra parameters to set as attributes on the sampler object\n :param dot_graph: str: A string specifying the causal graph.\n :param common_causes: list: A list of strings containing the variable names to control for.\n :param estimand_type: str: 'nonparametric-ate' is the only one currently supported. Others may be added later, to allow for specific, parametric estimands.\n :param proceed_when_unidentifiable: bool: A flag to over-ride user prompts to proceed when effects aren't\n identifiable with the assumptions provided.\n :param stateful: bool: Whether to retain state. By default, the do operation is stateless.\n :return: pandas.DataFrame: A DataFrame containing the sampled outcome\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d002d1911d9b6ad53cf0adca340ece83cdf4c874 | 81,309 | ipynb | Jupyter Notebook | use_case/01_intro_tutorial.ipynb | elekt/datenguide-python | 2f764c0a56500a95bf1829684ad96cdcae571037 | [
"MIT"
] | 1 | 2020-07-15T17:06:43.000Z | 2020-07-15T17:06:43.000Z | use_case/01_intro_tutorial.ipynb | elekt/datenguide-python | 2f764c0a56500a95bf1829684ad96cdcae571037 | [
"MIT"
] | null | null | null | use_case/01_intro_tutorial.ipynb | elekt/datenguide-python | 2f764c0a56500a95bf1829684ad96cdcae571037 | [
"MIT"
] | null | null | null | 34.540782 | 269 | 0.409241 | [
[
[
"# Welcome to the Datenguide Python Package\n\nWithin this notebook the functionality of the package will be explained and demonstrated with examples.\n\n### Topics\n\n- Import\n- get region IDs\n- get statstic IDs\n- get the data\n - for single regions\n - for multiple regions",
"_____no_output_____"
],
[
"## 1. Import",
"_____no_output_____"
],
[
"**Import the helper functions 'get_all_regions' and 'get_statistics'**\n\n**Import the module Query for the main functionality**",
"_____no_output_____"
]
],
[
[
"# ONLY FOR TESTING LOCAL PACKAGE\n# %cd ..\n\nfrom datenguidepy.query_helper import get_all_regions, get_statistics\nfrom datenguidepy import Query",
"C:\\Users\\Alexandra\\Documents\\GitHub\\datenguide-python\n"
]
],
[
[
"**Import pandas and matplotlib for the usual display of data as tables and graphs**",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib\n%matplotlib inline\n\npd.set_option('display.max_colwidth', 150)",
"_____no_output_____"
]
],
[
[
"## 2. Get Region IDs\n### How to get the ID of the region I want to query",
"_____no_output_____"
],
[
"Regionalstatistik - the database behind Datenguide - has data for differently granular levels of Germany. \n\nnuts:\n\n 1 – Bundesländer\n 2 – Regierungsbezirke / statistische Regionen\n 3 – Kreise / kreisfreie Städte.\n \nlau:\n\n 1 - Verwaltungsgemeinschaften\n 2 - Gemeinden.\n\nthe function `get_all_regions()` returns all IDs from all levels.",
"_____no_output_____"
]
],
[
[
"# get_all_regions returns all ids\nget_all_regions()",
"_____no_output_____"
]
],
[
[
"To get a specific ID, use the common pandas function `query()`\n",
"_____no_output_____"
]
],
[
[
"# e.g. get all \"Bundesländer\nget_all_regions().query(\"level == 'nuts1'\")",
"_____no_output_____"
],
[
"# e.g. get the ID of Havelland\nget_all_regions().query(\"name =='Havelland'\")",
"_____no_output_____"
]
],
[
[
"## 3. Get statistic IDs\n### How to find statistics",
"_____no_output_____"
]
],
[
[
"# get all statistics\nget_statistics()",
"_____no_output_____"
]
],
[
[
"If you already know the statsitic ID you are looking for - perfect. \n\nOtherwise you can use the pandas `query()` function so search e.g. for specific terms.",
"_____no_output_____"
]
],
[
[
"# find out the name of the desired statistic about birth\nget_statistics().query('long_description.str.contains(\"Statistik der Geburten\")', engine='python')",
"_____no_output_____"
]
],
[
[
"## 4. get the data",
"_____no_output_____"
],
[
"The top level element is the Query. For each query fields can be added (usually statistics / measures) that you want to get information on.",
"_____no_output_____"
],
[
"A Query can either be done on a single region, or on multiple regions (e.g. all Bundesländer).",
"_____no_output_____"
],
[
"### Single Region\n\nIf I want information - e.g. all births for the past years in Berlin:",
"_____no_output_____"
]
],
[
[
"# create a query for the region 11\nquery = Query.region('11')",
"_____no_output_____"
],
[
"# add a field (the statstic) to the query\nfield_births = query.add_field('BEV001')",
"_____no_output_____"
],
[
"# get the data of this query\nquery.results().head()",
"_____no_output_____"
]
],
[
[
"To get the short description in the result data frame instead of the cryptic ID (e.g. \"Lebend Geborene\" instead of BEV001) set the argument \"verbose_statsitics\"=True in the resutls:",
"_____no_output_____"
]
],
[
[
"query.results(verbose_statistics =True).head()",
"_____no_output_____"
]
],
[
[
"Now we only get the information about the count of births per year and the source of the data (year, value and source are default fields).\nBut there is more information in the statistic that we can get information on.\n\nLet's look at the meta data of the statstic:",
"_____no_output_____"
]
],
[
[
"# get information on the field\nfield_births.get_info()",
"\u001b[1mkind:\u001b[0m\nOBJECT\n\n\u001b[1mdescription:\u001b[0m\nLebend Geborene\n\n\u001b[1marguments:\u001b[0m\n\u001b[4myear\u001b[0m: LIST of type SCALAR(Int)\n\n\u001b[4mstatistics\u001b[0m: LIST of type ENUM(BEV001Statistics)\nenum values:\nR12612: Statistik der Geburten\n\n\u001b[4mALTMT1\u001b[0m: LIST of type ENUM(ALTMT1)\nenum values:\nALT000B20: unter 20 Jahre\nALT020B25: 20 bis unter 25 Jahre\nALT025B30: 25 bis unter 30 Jahre\nALT030B35: 30 bis unter 35 Jahre\nALT035B40: 35 bis unter 40 Jahre\nALT040UM: 40 Jahre und mehr\nGESAMT: Gesamt\n\n\u001b[4mGES\u001b[0m: LIST of type ENUM(GES)\nenum values:\nGESM: männlich\nGESW: weiblich\nGESAMT: Gesamt\n\n\u001b[4mNATEL1\u001b[0m: LIST of type ENUM(NATEL1)\nenum values:\nNATAAO: Mutter und Vater Ausländer, ohne Angabe der Nationalität\nNATDDDO: Mutter und Vater Deutsche, Mutter Deutsche und Vater o.Angabe der Nat.\nNATEETA: ein Elternteil Ausländer\nGESAMT: Gesamt\n\n\u001b[4mNAT\u001b[0m: LIST of type ENUM(NAT)\nenum values:\nNATA: Ausländer(innen)\nNATD: Deutsche\nGESAMT: Gesamt\n\n\u001b[4mLEGIT2\u001b[0m: LIST of type ENUM(LEGIT2)\nenum values:\nLEGIT01A: Eltern miteinander verheiratet\nLEGIT02A: Eltern nicht miteinander verheiratet\nGESAMT: Gesamt\n\n\u001b[4mBEVM01\u001b[0m: LIST of type ENUM(BEVM01)\nenum values:\nMONAT01: Januar\nMONAT02: Februar\nMONAT03: März\nMONAT04: April\nMONAT05: Mai\nMONAT06: Juni\nMONAT07: Juli\nMONAT08: August\nMONAT09: September\nMONAT10: Oktober\nMONAT11: November\nMONAT12: Dezember\nGESAMT: Gesamt\n\n\u001b[4mfilter\u001b[0m: INPUT_OBJECT(BEV001Filter)\n\n\u001b[1mfields:\u001b[0m\nid: Interne eindeutige ID\nyear: Jahr des Stichtages\nvalue: Wert\nsource: Quellenverweis zur GENESIS Regionaldatenbank\nALTMT1: Altersgruppen der Mutter (unter 20 bis 40 u.m.)\nGES: Geschlecht\nNATEL1: Nationalität der Eltern\nNAT: Nationalität\nLEGIT2: Legitimität\nBEVM01: Monat der Geburt\n\n\u001b[1menum values:\u001b[0m\nNone\n"
]
],
[
[
"The arguments tell us what we can use for filtering (e.g. only data on baby girls (female)).\n\nThe fields tell us what more information can be displayed in our results. ",
"_____no_output_____"
]
],
[
[
"# add filter\nfield_births.add_args({'GES': 'GESW'})",
"_____no_output_____"
],
[
"# now only about half the amount of births are returned as only the results for female babies are queried\nquery.results().head()",
"_____no_output_____"
],
[
"# add the field NAT (nationality) to the results\nfield_births.add_field('NAT')",
"_____no_output_____"
]
],
[
[
"**CAREFUL**: The information for the fields (e.g. nationality) is by default returned as a total amount. Therefore - if no argument \"NAT\" is specified in addition to the field, then only \"None\" will be displayed.\n\nIn order to get information on all possible values, the argument \"ALL\" needs to be added:\n(the rows with value \"None\" are the aggregated values of all options)",
"_____no_output_____"
]
],
[
[
"field_births.add_args({'NAT': 'ALL'})",
"_____no_output_____"
],
[
"query.results().head()",
"_____no_output_____"
]
],
[
[
"To display the short description of the enum values instead of the cryptic IDs (e.g. Ausländer(innen) instead of NATA), set the argument \"verbose_enums = True\" on the results:",
"_____no_output_____"
]
],
[
[
"query.results(verbose_enums=True).head()",
"_____no_output_____"
]
],
[
[
"## Multiple Regions",
"_____no_output_____"
],
[
"To display data for multiple single regions, a list with region IDs can be used:",
"_____no_output_____"
]
],
[
[
"query_multiple = Query.region(['01', '02'])\nquery_multiple.add_field('BEV001')\nquery_multiple.results().sort_values('year').head()",
"_____no_output_____"
]
],
[
[
"To display data for e.g. all 'Bundesländer' or for all regions within a Bundesland, you can use the function `all_regions()`:\n\n- specify nuts level\n- specify lau level\n- specify parent ID (Careful: not only the regions for the next lower level will be returned, but all levels - e.g. if you specify a parent on nuts level 1 then the \"children\" on nuts 2 but also the \"grandchildren\" on nuts 3, lau 1 and lau 2 will be returned)",
"_____no_output_____"
]
],
[
[
"# get data for all Bundesländer\nquery_all = Query.all_regions(nuts=1)\nquery_all.add_field('BEV001')\nquery_all.results().sort_values('year').head(12)",
"_____no_output_____"
],
[
"# get data for all regions within Brandenburg\nquery_all = Query.all_regions(parent='12')\nquery_all.add_field('BEV001')\nquery_all.results().head()",
"_____no_output_____"
],
[
"# get data for all nuts 3 regions within Brandenburg\nquery_all = Query.all_regions(parent='12', nuts=3)\nquery_all.add_field('BEV001')\nquery_all.results().sort_values('year').head()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d002e156d97ada6b864f67260dc27a30bd6903b6 | 10,991 | ipynb | Jupyter Notebook | dadosBQ_Pandas.ipynb | DrumondVilela/ProjetoFinal | 61887cfdc4284083237e82f168be4f78732d394f | [
"MIT"
] | null | null | null | dadosBQ_Pandas.ipynb | DrumondVilela/ProjetoFinal | 61887cfdc4284083237e82f168be4f78732d394f | [
"MIT"
] | null | null | null | dadosBQ_Pandas.ipynb | DrumondVilela/ProjetoFinal | 61887cfdc4284083237e82f168be4f78732d394f | [
"MIT"
] | null | null | null | 28.400517 | 152 | 0.472386 | [
[
[
"pip install pandera",
"_____no_output_____"
],
[
"pip install gcsfs",
"_____no_output_____"
],
[
"import os\nimport pandas as pd\nfrom google.cloud import storage",
"_____no_output_____"
],
[
"serviceAccount = '/content/Chave Ingestao Apache.json'\nos.environ['GOOGLE_APPLICATION_CREDENTIALS'] = serviceAccount",
"_____no_output_____"
],
[
"#leitura do arquivo em JSON\ndf = pd.read_json(r'gs://projeto-final-grupo09/entrada_dados/Projeto Final', lines = True)\ndf.head(1)",
"_____no_output_____"
],
[
"#Renomeando colunas\ndf.rename(columns={'id':'identificacao','created_on':'criado_em','operation':'operacao','property_type':'tipo_propriedade',\n 'place_name':'nome_do_local','place_with_parent_names':'pais_local','country_name':'pais','state_name':'estado',\n 'geonames_id':'g_nomes','lat_lon':'latitude_longitude','lat':'latitude','lon':'longitude','price':\n 'preco_cheio','currency':'moeda','price_aprox_local_currency':'preco',\n 'price_aprox_usd':'preco_aproximado_dolar','surface_total_in_m2':'area_total_por_m2',\n 'surface_covered_in_m2':'area_construcao_em_m2','price_usd_per_m2':'preco_dolar_por_m2',\n 'price_per_m2':'preco_por_m2','floor':'andar','rooms':'quartos','expenses':'despesas',\n 'properati_url':'url_da_propriedade','description':'descricao', 'title':'titulo',\n 'image_thumbnail':'miniatura_imagem'}, inplace = True)\ndf.head(2)\n",
"_____no_output_____"
],
[
"#chamar a coluna de operacao para ver se tem algo além de venda (no caso a coluna só tem sell - venda - entao irei dropar posteriormente)\nsorted(pd.unique(df['operacao']))",
"_____no_output_____"
],
[
"#chamar a coluna de país para ver se tem algo além de Brasil (no caso só tem Brasil - então iremos dropar posteriormente)\nsorted(pd.unique(df['pais']))",
"_____no_output_____"
],
[
"#chamar a coluna de moeda para ver se tem algo além de BRL (no caso só tem BRL - então iremos dropar posteriormente)\nsorted(pd.unique(df['moeda']))",
"_____no_output_____"
],
[
"#criacao de variavel - colunas - para posterior drop\ncolunas = ['operacao', 'pais', 'moeda', 'latitude_longitude', 'latitude', 'longitude', 'preco_aproximado_dolar', 'pais_local', \n'preco_dolar_por_m2', 'andar', 'despesas', 'descricao', 'titulo', 'miniatura_imagem', 'url_da_propriedade', 'preco_cheio']\ndf.drop(colunas, axis=1, inplace=True)",
"_____no_output_____"
],
[
"#verificando se há (e quantos são) os valores na coluna nome_do_local\ndf['nome_do_local'].value_counts()",
"_____no_output_____"
],
[
"#verificar se há apenas um valor na coluna (no caso a coluna propriedade_tipo tem 3 informações significativas e uma (PH) que será dropada)\nsorted(pd.unique(df['tipo_propriedade']))",
"_____no_output_____"
],
[
"#contando quantos valores tem cada um dos itens em tipo_propriedade - casa, apartamento e lojas\ndf['tipo_propriedade'].value_counts()",
"_____no_output_____"
],
[
"#traduzindo as informações contidas na coluna de tipo_propriedade\ndf['tipo_propriedade'].replace(['house', 'apartment', 'store'],['casa','apartamento','loja'], inplace = True)",
"_____no_output_____"
],
[
"#quantidade de quartos e salas (no caso das lojas)\ndf['quartos'].value_counts()",
"_____no_output_____"
],
[
"#chamar a coluna de quartos para descobrir quais os valores contidos\nsorted(pd.unique(df['quartos']))",
"_____no_output_____"
],
[
"#devido a coluna quartos ser um float, forçamos ele a se tornar um numero inteiro e o NaN se tornar 0\ndf['quartos'] = df['quartos'].fillna(0.0).astype(int)",
"_____no_output_____"
],
[
"df.head(10)",
"_____no_output_____"
],
[
"df.to_csv(\"gs://lucao-buck\", sep=\",\", index=False)",
"_____no_output_____"
],
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d002e9694740606d508ae0ab95f60cdabad6c231 | 1,949 | ipynb | Jupyter Notebook | python-challenges/Challenges/.ipynb_checkpoints/Invert-binary-tree-checkpoint.ipynb | coopersec/research-learning | da65e7999f0f9948c85dc2a74d15b25dbc3f7108 | [
"MIT"
] | 1 | 2022-02-10T23:59:46.000Z | 2022-02-10T23:59:46.000Z | python-challenges/Challenges/Invert-binary-tree.ipynb | coopersec/research-learning | da65e7999f0f9948c85dc2a74d15b25dbc3f7108 | [
"MIT"
] | null | null | null | python-challenges/Challenges/Invert-binary-tree.ipynb | coopersec/research-learning | da65e7999f0f9948c85dc2a74d15b25dbc3f7108 | [
"MIT"
] | null | null | null | 22.929412 | 97 | 0.471524 | [
[
[
"# Definition for a binary tree node.\nclass Node:\n def __init__(self, data, val=0, left=None, right=None):\n self.data = data\n self.val = val\n self.left = left\n self.right = right\nclass Solution:\n def invertTree(self, root: Node) -> Node:\n if root is None:\n return None\n \n print(root.data, end=' ')\n root.left\n root.right\n self.invertTree(root.right) \n self.invertTree(root.left)\n #root.left, root.right = self.invertTree(root.right), self.invertTree(root.left)\n return root\nroot = Node(1)\nroot.left = Node(2)\nroot.right = Node(3)\nroot.left.left = Node(4)\nroot.left.right = Node(5)\nroot.right.left = Node(6)\nroot.right.right = Node(7)\n\na = Solution()\na.invertTree(root)\n\n\n \n ",
"1 3 7 6 2 5 4 "
]
]
] | [
"code"
] | [
[
"code"
]
] |
d002ec87edea457693b0b71f1c924cd62b4f7937 | 5,646 | ipynb | Jupyter Notebook | notebooks/ch03.ipynb | jkurdys/ThinkPython2 | 7bdbe11f6ef62eac29ee7d06170bd734a061cb0b | [
"MIT"
] | null | null | null | notebooks/ch03.ipynb | jkurdys/ThinkPython2 | 7bdbe11f6ef62eac29ee7d06170bd734a061cb0b | [
"MIT"
] | null | null | null | notebooks/ch03.ipynb | jkurdys/ThinkPython2 | 7bdbe11f6ef62eac29ee7d06170bd734a061cb0b | [
"MIT"
] | null | null | null | 18.45098 | 80 | 0.374956 | [
[
[
"def repeat_lyrics():\n print_lyrics()\n print_lyrics()",
"_____no_output_____"
],
[
"def print_lyrics():\n print('hi')\n print('how do you do?')\n ",
"_____no_output_____"
],
[
"repeat_lyrics()",
"hi\nhow do you do?\nhi\nhow do you do?\n"
],
[
"def print_twice(bruce):\n print(bruce)\n print(bruce)\n ",
"_____no_output_____"
],
[
"print_twice((bruce + ' ')*4)",
"twice twice twice twice \ntwice twice twice twice \n"
],
[
"bruce = 'twice'",
"_____no_output_____"
],
[
"def right_justify(s):\n print(((70 - len(s)) * ' ') + s)",
"_____no_output_____"
],
[
"right_justify('am I right?')",
" am I right?\n"
],
[
"right_justify('no, I am!')",
" no, I am!\n"
],
[
"def do_twice(f, v):\n f(v)\n f(v)",
"_____no_output_____"
],
[
"def print_spam():\n print('spam')",
"_____no_output_____"
],
[
"do_twice(print_twice, 'spam')",
"spam\nspam\nspam\nspam\n"
],
[
"def do_four(f, v):\n do_twice(f, v)\n do_twice(f, v)",
"_____no_output_____"
],
[
"do_four(print_twice, 'spam')",
"spam\nspam\nspam\nspam\nspam\nspam\nspam\nspam\n"
],
[
"print(2 * ('+ - - - - ') + '+')",
"+ - - - - + - - - - +\n"
],
[
"print(2 * ('+ - - - - ') + '+')\nprint(2 * ('| ') + '|')\nprint(2 * ('| ') + '|')\nprint(2 * ('| ') + '|')\nprint(2 * ('| ') + '|')\nprint(2 * ('+ - - - - ') + '+')\nprint(2 * ('| ') + '|')\nprint(2 * ('| ') + '|')\nprint(2 * ('| ') + '|')\nprint(2 * ('| ') + '|')\nprint(2 * ('+ - - - - ') + '+')",
"+ - - - - + - - - - +\n| | |\n| | |\n| | |\n| | |\n+ - - - - + - - - - +\n| | |\n| | |\n| | |\n| | |\n+ - - - - + - - - - +\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d002f51b520dfb6f7f7c8f13e0401f22dc925760 | 633,929 | ipynb | Jupyter Notebook | Chapter4_TheGreatestTheoremNeverTold/Ch4_LawOfLargeNumbers_PyMC3.ipynb | quantopian/Probabilistic-Programming-and-Bayesian-Methods-for-Hackers | 11006cea89d6b2cbf4fc06173d717d3f08966f93 | [
"MIT"
] | 74 | 2016-07-22T19:03:32.000Z | 2022-03-24T04:23:28.000Z | Chapter4_TheGreatestTheoremNeverTold/Ch4_LawOfLargeNumbers_PyMC3.ipynb | noisyoscillator/Probabilistic-Programming-and-Bayesian-Methods-for-Hackers | 11006cea89d6b2cbf4fc06173d717d3f08966f93 | [
"MIT"
] | 7 | 2016-08-02T08:17:15.000Z | 2016-10-03T21:48:59.000Z | Chapter4_TheGreatestTheoremNeverTold/Ch4_LawOfLargeNumbers_PyMC3.ipynb | noisyoscillator/Probabilistic-Programming-and-Bayesian-Methods-for-Hackers | 11006cea89d6b2cbf4fc06173d717d3f08966f93 | [
"MIT"
] | 39 | 2016-07-23T01:42:20.000Z | 2022-02-11T14:55:26.000Z | 528.274167 | 117,692 | 0.919833 | [
[
[
"# Chapter 4\n`Original content created by Cam Davidson-Pilon`\n\n`Ported to Python 3 and PyMC3 by Max Margenot (@clean_utensils) and Thomas Wiecki (@twiecki) at Quantopian (@quantopian)`\n\n______\n\n## The greatest theorem never told\n\n\nThis chapter focuses on an idea that is always bouncing around our minds, but is rarely made explicit outside books devoted to statistics. In fact, we've been using this simple idea in every example thus far. ",
"_____no_output_____"
],
[
"### The Law of Large Numbers\n\nLet $Z_i$ be $N$ independent samples from some probability distribution. According to *the Law of Large numbers*, so long as the expected value $E[Z]$ is finite, the following holds,\n\n$$\\frac{1}{N} \\sum_{i=1}^N Z_i \\rightarrow E[ Z ], \\;\\;\\; N \\rightarrow \\infty.$$\n\nIn words:\n\n> The average of a sequence of random variables from the same distribution converges to the expected value of that distribution.\n\nThis may seem like a boring result, but it will be the most useful tool you use.",
"_____no_output_____"
],
[
"### Intuition \n\nIf the above Law is somewhat surprising, it can be made more clear by examining a simple example. \n\nConsider a random variable $Z$ that can take only two values, $c_1$ and $c_2$. Suppose we have a large number of samples of $Z$, denoting a specific sample $Z_i$. The Law says that we can approximate the expected value of $Z$ by averaging over all samples. Consider the average:\n\n\n$$ \\frac{1}{N} \\sum_{i=1}^N \\;Z_i $$\n\n\nBy construction, $Z_i$ can only take on $c_1$ or $c_2$, hence we can partition the sum over these two values:\n\n\\begin{align}\n\\frac{1}{N} \\sum_{i=1}^N \\;Z_i\n& =\\frac{1}{N} \\big( \\sum_{ Z_i = c_1}c_1 + \\sum_{Z_i=c_2}c_2 \\big) \\\\\\\\[5pt]\n& = c_1 \\sum_{ Z_i = c_1}\\frac{1}{N} + c_2 \\sum_{ Z_i = c_2}\\frac{1}{N} \\\\\\\\[5pt]\n& = c_1 \\times \\text{ (approximate frequency of $c_1$) } \\\\\\\\ \n& \\;\\;\\;\\;\\;\\;\\;\\;\\; + c_2 \\times \\text{ (approximate frequency of $c_2$) } \\\\\\\\[5pt]\n& \\approx c_1 \\times P(Z = c_1) + c_2 \\times P(Z = c_2 ) \\\\\\\\[5pt]\n& = E[Z]\n\\end{align}\n\n\nEquality holds in the limit, but we can get closer and closer by using more and more samples in the average. This Law holds for almost *any distribution*, minus some important cases we will encounter later.\n\n##### Example\n____\n\n\nBelow is a diagram of the Law of Large numbers in action for three different sequences of Poisson random variables. \n\n We sample `sample_size = 100000` Poisson random variables with parameter $\\lambda = 4.5$. (Recall the expected value of a Poisson random variable is equal to it's parameter.) We calculate the average for the first $n$ samples, for $n=1$ to `sample_size`. ",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nfrom IPython.core.pylabtools import figsize\nimport matplotlib.pyplot as plt\n\nfigsize( 12.5, 5 )\n\nsample_size = 100000\nexpected_value = lambda_ = 4.5\npoi = np.random.poisson\nN_samples = range(1,sample_size,100)\n\nfor k in range(3):\n\n samples = poi( lambda_, sample_size ) \n \n partial_average = [ samples[:i].mean() for i in N_samples ]\n \n plt.plot( N_samples, partial_average, lw=1.5,label=\"average \\\nof $n$ samples; seq. %d\"%k)\n \n\nplt.plot( N_samples, expected_value*np.ones_like( partial_average), \n ls = \"--\", label = \"true expected value\", c = \"k\" )\n\nplt.ylim( 4.35, 4.65) \nplt.title( \"Convergence of the average of \\n random variables to its \\\nexpected value\" )\nplt.ylabel( \"average of $n$ samples\" )\nplt.xlabel( \"# of samples, $n$\")\nplt.legend();",
"_____no_output_____"
]
],
[
[
"Looking at the above plot, it is clear that when the sample size is small, there is greater variation in the average (compare how *jagged and jumpy* the average is initially, then *smooths* out). All three paths *approach* the value 4.5, but just flirt with it as $N$ gets large. Mathematicians and statistician have another name for *flirting*: convergence. \n\nAnother very relevant question we can ask is *how quickly am I converging to the expected value?* Let's plot something new. For a specific $N$, let's do the above trials thousands of times and compute how far away we are from the true expected value, on average. But wait — *compute on average*? This is simply the law of large numbers again! For example, we are interested in, for a specific $N$, the quantity:\n\n$$D(N) = \\sqrt{ \\;E\\left[\\;\\; \\left( \\frac{1}{N}\\sum_{i=1}^NZ_i - 4.5 \\;\\right)^2 \\;\\;\\right] \\;\\;}$$\n\nThe above formulae is interpretable as a distance away from the true value (on average), for some $N$. (We take the square root so the dimensions of the above quantity and our random variables are the same). As the above is an expected value, it can be approximated using the law of large numbers: instead of averaging $Z_i$, we calculate the following multiple times and average them:\n\n$$ Y_k = \\left( \\;\\frac{1}{N}\\sum_{i=1}^NZ_i - 4.5 \\; \\right)^2 $$\n\nBy computing the above many, $N_y$, times (remember, it is random), and averaging them:\n\n$$ \\frac{1}{N_Y} \\sum_{k=1}^{N_Y} Y_k \\rightarrow E[ Y_k ] = E\\;\\left[\\;\\; \\left( \\frac{1}{N}\\sum_{i=1}^NZ_i - 4.5 \\;\\right)^2 \\right]$$\n\nFinally, taking the square root:\n\n$$ \\sqrt{\\frac{1}{N_Y} \\sum_{k=1}^{N_Y} Y_k} \\approx D(N) $$ ",
"_____no_output_____"
]
],
[
[
"figsize( 12.5, 4)\n\nN_Y = 250 #use this many to approximate D(N)\nN_array = np.arange( 1000, 50000, 2500 ) #use this many samples in the approx. to the variance.\nD_N_results = np.zeros( len( N_array ) )\n\nlambda_ = 4.5 \nexpected_value = lambda_ #for X ~ Poi(lambda) , E[ X ] = lambda\n\ndef D_N( n ):\n \"\"\"\n This function approx. D_n, the average variance of using n samples.\n \"\"\"\n Z = poi( lambda_, (n, N_Y) )\n average_Z = Z.mean(axis=0)\n return np.sqrt( ( (average_Z - expected_value)**2 ).mean() )\n \n \nfor i,n in enumerate(N_array):\n D_N_results[i] = D_N(n)\n\n\nplt.xlabel( \"$N$\" )\nplt.ylabel( \"expected squared-distance from true value\" )\nplt.plot(N_array, D_N_results, lw = 3, \n label=\"expected distance between\\n\\\nexpected value and \\naverage of $N$ random variables.\")\nplt.plot( N_array, np.sqrt(expected_value)/np.sqrt(N_array), lw = 2, ls = \"--\", \n label = r\"$\\frac{\\sqrt{\\lambda}}{\\sqrt{N}}$\" )\nplt.legend()\nplt.title( \"How 'fast' is the sample average converging? \" );",
"_____no_output_____"
]
],
[
[
"As expected, the expected distance between our sample average and the actual expected value shrinks as $N$ grows large. But also notice that the *rate* of convergence decreases, that is, we need only 10 000 additional samples to move from 0.020 to 0.015, a difference of 0.005, but *20 000* more samples to again decrease from 0.015 to 0.010, again only a 0.005 decrease.\n\n\nIt turns out we can measure this rate of convergence. Above I have plotted a second line, the function $\\sqrt{\\lambda}/\\sqrt{N}$. This was not chosen arbitrarily. In most cases, given a sequence of random variable distributed like $Z$, the rate of convergence to $E[Z]$ of the Law of Large Numbers is \n\n$$ \\frac{ \\sqrt{ \\; Var(Z) \\; } }{\\sqrt{N} }$$\n\nThis is useful to know: for a given large $N$, we know (on average) how far away we are from the estimate. On the other hand, in a Bayesian setting, this can seem like a useless result: Bayesian analysis is OK with uncertainty so what's the *statistical* point of adding extra precise digits? Though drawing samples can be so computationally cheap that having a *larger* $N$ is fine too. \n\n### How do we compute $Var(Z)$ though?\n\nThe variance is simply another expected value that can be approximated! Consider the following, once we have the expected value (by using the Law of Large Numbers to estimate it, denote it $\\mu$), we can estimate the variance:\n\n$$ \\frac{1}{N}\\sum_{i=1}^N \\;(Z_i - \\mu)^2 \\rightarrow E[ \\;( Z - \\mu)^2 \\;] = Var( Z )$$\n\n### Expected values and probabilities \nThere is an even less explicit relationship between expected value and estimating probabilities. Define the *indicator function*\n\n$$\\mathbb{1}_A(x) = \n\\begin{cases} 1 & x \\in A \\\\\\\\\n 0 & else\n\\end{cases}\n$$\nThen, by the law of large numbers, if we have many samples $X_i$, we can estimate the probability of an event $A$, denoted $P(A)$, by:\n\n$$ \\frac{1}{N} \\sum_{i=1}^N \\mathbb{1}_A(X_i) \\rightarrow E[\\mathbb{1}_A(X)] = P(A) $$\n\nAgain, this is fairly obvious after a moments thought: the indicator function is only 1 if the event occurs, so we are summing only the times the event occurs and dividing by the total number of trials (consider how we usually approximate probabilities using frequencies). For example, suppose we wish to estimate the probability that a $Z \\sim Exp(.5)$ is greater than 5, and we have many samples from a $Exp(.5)$ distribution. \n\n\n$$ P( Z > 5 ) = \\sum_{i=1}^N \\mathbb{1}_{z > 5 }(Z_i) $$\n",
"_____no_output_____"
]
],
[
[
"N = 10000\nprint( np.mean( [ np.random.exponential( 0.5 ) > 5 for i in range(N) ] ) )",
"0.0001\n"
]
],
[
[
"### What does this all have to do with Bayesian statistics? \n\n\n*Point estimates*, to be introduced in the next chapter, in Bayesian inference are computed using expected values. In more analytical Bayesian inference, we would have been required to evaluate complicated expected values represented as multi-dimensional integrals. No longer. If we can sample from the posterior distribution directly, we simply need to evaluate averages. Much easier. If accuracy is a priority, plots like the ones above show how fast you are converging. And if further accuracy is desired, just take more samples from the posterior. \n\nWhen is enough enough? When can you stop drawing samples from the posterior? That is the practitioners decision, and also dependent on the variance of the samples (recall from above a high variance means the average will converge slower). \n\nWe also should understand when the Law of Large Numbers fails. As the name implies, and comparing the graphs above for small $N$, the Law is only true for large sample sizes. Without this, the asymptotic result is not reliable. Knowing in what situations the Law fails can give us *confidence in how unconfident we should be*. The next section deals with this issue.",
"_____no_output_____"
],
[
"## The Disorder of Small Numbers\n\nThe Law of Large Numbers is only valid as $N$ gets *infinitely* large: never truly attainable. While the law is a powerful tool, it is foolhardy to apply it liberally. Our next example illustrates this.\n\n\n##### Example: Aggregated geographic data\n\n\nOften data comes in aggregated form. For instance, data may be grouped by state, county, or city level. Of course, the population numbers vary per geographic area. If the data is an average of some characteristic of each the geographic areas, we must be conscious of the Law of Large Numbers and how it can *fail* for areas with small populations.\n\nWe will observe this on a toy dataset. Suppose there are five thousand counties in our dataset. Furthermore, population number in each state are uniformly distributed between 100 and 1500. The way the population numbers are generated is irrelevant to the discussion, so we do not justify this. We are interested in measuring the average height of individuals per county. Unbeknownst to us, height does **not** vary across county, and each individual, regardless of the county he or she is currently living in, has the same distribution of what their height may be:\n\n$$ \\text{height} \\sim \\text{Normal}(150, 15 ) $$\n\nWe aggregate the individuals at the county level, so we only have data for the *average in the county*. What might our dataset look like?",
"_____no_output_____"
]
],
[
[
"figsize( 12.5, 4) \nstd_height = 15\nmean_height = 150\n\nn_counties = 5000\npop_generator = np.random.randint\nnorm = np.random.normal\n\n#generate some artificial population numbers\npopulation = pop_generator(100, 1500, n_counties )\n\naverage_across_county = np.zeros( n_counties )\nfor i in range( n_counties ):\n #generate some individuals and take the mean\n average_across_county[i] = norm(mean_height, 1./std_height,\n population[i] ).mean()\n \n#located the counties with the apparently most extreme average heights.\ni_min = np.argmin( average_across_county )\ni_max = np.argmax( average_across_county )\n\n#plot population size vs. recorded average\nplt.scatter( population, average_across_county, alpha = 0.5, c=\"#7A68A6\")\nplt.scatter( [ population[i_min], population[i_max] ], \n [average_across_county[i_min], average_across_county[i_max] ],\n s = 60, marker = \"o\", facecolors = \"none\",\n edgecolors = \"#A60628\", linewidths = 1.5, \n label=\"extreme heights\")\n\nplt.xlim( 100, 1500 )\nplt.title( \"Average height vs. County Population\")\nplt.xlabel(\"County Population\")\nplt.ylabel(\"Average height in county\")\nplt.plot( [100, 1500], [150, 150], color = \"k\", label = \"true expected \\\nheight\", ls=\"--\" )\nplt.legend(scatterpoints = 1);",
"_____no_output_____"
]
],
[
[
"What do we observe? *Without accounting for population sizes* we run the risk of making an enormous inference error: if we ignored population size, we would say that the county with the shortest and tallest individuals have been correctly circled. But this inference is wrong for the following reason. These two counties do *not* necessarily have the most extreme heights. The error results from the calculated average of smaller populations not being a good reflection of the true expected value of the population (which in truth should be $\\mu =150$). The sample size/population size/$N$, whatever you wish to call it, is simply too small to invoke the Law of Large Numbers effectively. \n\nWe provide more damning evidence against this inference. Recall the population numbers were uniformly distributed over 100 to 1500. Our intuition should tell us that the counties with the most extreme population heights should also be uniformly spread over 100 to 4000, and certainly independent of the county's population. Not so. Below are the population sizes of the counties with the most extreme heights.",
"_____no_output_____"
]
],
[
[
"print(\"Population sizes of 10 'shortest' counties: \")\nprint(population[ np.argsort( average_across_county )[:10] ], '\\n')\nprint(\"Population sizes of 10 'tallest' counties: \")\nprint(population[ np.argsort( -average_across_county )[:10] ])",
"Population sizes of 10 'shortest' counties: \n[109 135 135 133 109 157 175 120 105 131] \n\nPopulation sizes of 10 'tallest' counties: \n[122 133 313 109 124 280 106 198 326 216]\n"
]
],
[
[
"Not at all uniform over 100 to 1500. This is an absolute failure of the Law of Large Numbers. \n\n##### Example: Kaggle's *U.S. Census Return Rate Challenge*\n\nBelow is data from the 2010 US census, which partitions populations beyond counties to the level of block groups (which are aggregates of city blocks or equivalents). The dataset is from a Kaggle machine learning competition some colleagues and I participated in. The objective was to predict the census letter mail-back rate of a group block, measured between 0 and 100, using census variables (median income, number of females in the block-group, number of trailer parks, average number of children etc.). Below we plot the census mail-back rate versus block group population:",
"_____no_output_____"
]
],
[
[
"figsize( 12.5, 6.5 )\ndata = np.genfromtxt( \"./data/census_data.csv\", skip_header=1, \n delimiter= \",\")\nplt.scatter( data[:,1], data[:,0], alpha = 0.5, c=\"#7A68A6\")\nplt.title(\"Census mail-back rate vs Population\")\nplt.ylabel(\"Mail-back rate\")\nplt.xlabel(\"population of block-group\")\nplt.xlim(-100, 15e3 )\nplt.ylim( -5, 105)\n\ni_min = np.argmin( data[:,0] )\ni_max = np.argmax( data[:,0] )\n \nplt.scatter( [ data[i_min,1], data[i_max, 1] ], \n [ data[i_min,0], data[i_max,0] ],\n s = 60, marker = \"o\", facecolors = \"none\",\n edgecolors = \"#A60628\", linewidths = 1.5, \n label=\"most extreme points\")\n\nplt.legend(scatterpoints = 1);",
"_____no_output_____"
]
],
[
[
"The above is a classic phenomenon in statistics. I say *classic* referring to the \"shape\" of the scatter plot above. It follows a classic triangular form, that tightens as we increase the sample size (as the Law of Large Numbers becomes more exact). \n\nI am perhaps overstressing the point and maybe I should have titled the book *\"You don't have big data problems!\"*, but here again is an example of the trouble with *small datasets*, not big ones. Simply, small datasets cannot be processed using the Law of Large Numbers. Compare with applying the Law without hassle to big datasets (ex. big data). I mentioned earlier that paradoxically big data prediction problems are solved by relatively simple algorithms. The paradox is partially resolved by understanding that the Law of Large Numbers creates solutions that are *stable*, i.e. adding or subtracting a few data points will not affect the solution much. On the other hand, adding or removing data points to a small dataset can create very different results. \n\nFor further reading on the hidden dangers of the Law of Large Numbers, I would highly recommend the excellent manuscript [The Most Dangerous Equation](http://nsm.uh.edu/~dgraur/niv/TheMostDangerousEquation.pdf). ",
"_____no_output_____"
],
[
"##### Example: How to order Reddit submissions\n\nYou may have disagreed with the original statement that the Law of Large numbers is known to everyone, but only implicitly in our subconscious decision making. Consider ratings on online products: how often do you trust an average 5-star rating if there is only 1 reviewer? 2 reviewers? 3 reviewers? We implicitly understand that with such few reviewers that the average rating is **not** a good reflection of the true value of the product.\n\nThis has created flaws in how we sort items, and more generally, how we compare items. Many people have realized that sorting online search results by their rating, whether the objects be books, videos, or online comments, return poor results. Often the seemingly top videos or comments have perfect ratings only from a few enthusiastic fans, and truly more quality videos or comments are hidden in later pages with *falsely-substandard* ratings of around 4.8. How can we correct this?\n\nConsider the popular site Reddit (I purposefully did not link to the website as you would never come back). The site hosts links to stories or images, called submissions, for people to comment on. Redditors can vote up or down on each submission (called upvotes and downvotes). Reddit, by default, will sort submissions to a given subreddit by Hot, that is, the submissions that have the most upvotes recently.\n\n<img src=\"http://i.imgur.com/3v6bz9f.png\" />\n\n\nHow would you determine which submissions are the best? There are a number of ways to achieve this:\n\n1. *Popularity*: A submission is considered good if it has many upvotes. A problem with this model is that a submission with hundreds of upvotes, but thousands of downvotes. While being very *popular*, the submission is likely more controversial than best.\n2. *Difference*: Using the *difference* of upvotes and downvotes. This solves the above problem, but fails when we consider the temporal nature of submission. Depending on when a submission is posted, the website may be experiencing high or low traffic. The difference method will bias the *Top* submissions to be the those made during high traffic periods, which have accumulated more upvotes than submissions that were not so graced, but are not necessarily the best.\n3. *Time adjusted*: Consider using Difference divided by the age of the submission. This creates a *rate*, something like *difference per second*, or *per minute*. An immediate counter-example is, if we use per second, a 1 second old submission with 1 upvote would be better than a 100 second old submission with 99 upvotes. One can avoid this by only considering at least t second old submission. But what is a good t value? Does this mean no submission younger than t is good? We end up comparing unstable quantities with stable quantities (young vs. old submissions).\n3. *Ratio*: Rank submissions by the ratio of upvotes to total number of votes (upvotes plus downvotes). This solves the temporal issue, such that new submissions who score well can be considered Top just as likely as older submissions, provided they have many upvotes to total votes. The problem here is that a submission with a single upvote (ratio = 1.0) will beat a submission with 999 upvotes and 1 downvote (ratio = 0.999), but clearly the latter submission is *more likely* to be better.\n\nI used the phrase *more likely* for good reason. It is possible that the former submission, with a single upvote, is in fact a better submission than the later with 999 upvotes. The hesitation to agree with this is because we have not seen the other 999 potential votes the former submission might get. Perhaps it will achieve an additional 999 upvotes and 0 downvotes and be considered better than the latter, though not likely.\n\nWhat we really want is an estimate of the *true upvote ratio*. Note that the true upvote ratio is not the same as the observed upvote ratio: the true upvote ratio is hidden, and we only observe upvotes vs. downvotes (one can think of the true upvote ratio as \"what is the underlying probability someone gives this submission a upvote, versus a downvote\"). So the 999 upvote/1 downvote submission probably has a true upvote ratio close to 1, which we can assert with confidence thanks to the Law of Large Numbers, but on the other hand we are much less certain about the true upvote ratio of the submission with only a single upvote. Sounds like a Bayesian problem to me.\n\n",
"_____no_output_____"
],
[
"One way to determine a prior on the upvote ratio is to look at the historical distribution of upvote ratios. This can be accomplished by scraping Reddit's submissions and determining a distribution. There are a few problems with this technique though:\n\n1. Skewed data: The vast majority of submissions have very few votes, hence there will be many submissions with ratios near the extremes (see the \"triangular plot\" in the above Kaggle dataset), effectively skewing our distribution to the extremes. One could try to only use submissions with votes greater than some threshold. Again, problems are encountered. There is a tradeoff between number of submissions available to use and a higher threshold with associated ratio precision. \n2. Biased data: Reddit is composed of different subpages, called subreddits. Two examples are *r/aww*, which posts pics of cute animals, and *r/politics*. It is very likely that the user behaviour towards submissions of these two subreddits are very different: visitors are likely friendly and affectionate in the former, and would therefore upvote submissions more, compared to the latter, where submissions are likely to be controversial and disagreed upon. Therefore not all submissions are the same. \n\n\nIn light of these, I think it is better to use a `Uniform` prior.\n\n\nWith our prior in place, we can find the posterior of the true upvote ratio. The Python script `top_showerthoughts_submissions.py` will scrape the best posts from the `showerthoughts` community on Reddit. This is a text-only community so the title of each post *is* the post. Below is the top post as well as some other sample posts:",
"_____no_output_____"
]
],
[
[
"#adding a number to the end of the %run call with get the ith top post.\n%run top_showerthoughts_submissions.py 2\n\nprint(\"Post contents: \\n\")\nprint(top_post)",
"Post contents: \n\nToilet paper should be free and have advertising printed on it.\n"
],
[
"\"\"\"\ncontents: an array of the text from the last 100 top submissions to a subreddit\nvotes: a 2d numpy array of upvotes, downvotes for each submission.\n\"\"\"\nn_submissions = len(votes)\nsubmissions = np.random.randint( n_submissions, size=4)\nprint(\"Some Submissions (out of %d total) \\n-----------\"%n_submissions)\nfor i in submissions:\n print('\"' + contents[i] + '\"')\n print(\"upvotes/downvotes: \",votes[i,:], \"\\n\")",
"Some Submissions (out of 98 total) \n-----------\n\"Rappers from the 90's used guns when they had beef rappers today use Twitter.\"\nupvotes/downvotes: [32 3] \n\n\"All polls are biased towards people who are willing to take polls\"\nupvotes/downvotes: [1918 101] \n\n\"Taco Bell should give customers an extra tortilla so they can make a burrito out of all the stuff that spilled out of the other burritos they ate.\"\nupvotes/downvotes: [79 17] \n\n\"There should be an /r/alanismorissette where it's just examples of people using \"ironic\" incorrectly\"\nupvotes/downvotes: [33 6] \n\n"
]
],
[
[
" For a given true upvote ratio $p$ and $N$ votes, the number of upvotes will look like a Binomial random variable with parameters $p$ and $N$. (This is because of the equivalence between upvote ratio and probability of upvoting versus downvoting, out of $N$ possible votes/trials). We create a function that performs Bayesian inference on $p$, for a particular submission's upvote/downvote pair.",
"_____no_output_____"
]
],
[
[
"import pymc3 as pm\n\ndef posterior_upvote_ratio( upvotes, downvotes, samples = 20000):\n \"\"\"\n This function accepts the number of upvotes and downvotes a particular submission recieved, \n and the number of posterior samples to return to the user. Assumes a uniform prior.\n \"\"\"\n N = upvotes + downvotes\n with pm.Model() as model:\n upvote_ratio = pm.Uniform(\"upvote_ratio\", 0, 1)\n observations = pm.Binomial( \"obs\", N, upvote_ratio, observed=upvotes)\n \n trace = pm.sample(samples, step=pm.Metropolis())\n \n burned_trace = trace[int(samples/4):]\n return burned_trace[\"upvote_ratio\"]\n ",
"_____no_output_____"
]
],
[
[
"Below are the resulting posterior distributions.",
"_____no_output_____"
]
],
[
[
"figsize( 11., 8)\nposteriors = []\ncolours = [\"#348ABD\", \"#A60628\", \"#7A68A6\", \"#467821\", \"#CF4457\"]\nfor i in range(len(submissions)):\n j = submissions[i]\n posteriors.append( posterior_upvote_ratio( votes[j, 0], votes[j,1] ) )\n plt.hist( posteriors[i], bins = 10, normed = True, alpha = .9, \n histtype=\"step\",color = colours[i%5], lw = 3,\n label = '(%d up:%d down)\\n%s...'%(votes[j, 0], votes[j,1], contents[j][:50]) )\n plt.hist( posteriors[i], bins = 10, normed = True, alpha = .2, \n histtype=\"stepfilled\",color = colours[i], lw = 3, )\n \nplt.legend(loc=\"upper left\")\nplt.xlim( 0, 1)\nplt.title(\"Posterior distributions of upvote ratios on different submissions\");",
"Applied interval-transform to upvote_ratio and added transformed upvote_ratio_interval_ to model.\n [-------100%-------] 20000 of 20000 in 1.4 sec. | SPS: 14595.5 | ETA: 0.0Applied interval-transform to upvote_ratio and added transformed upvote_ratio_interval_ to model.\n [-------100%-------] 20000 of 20000 in 1.3 sec. | SPS: 15189.5 | ETA: 0.0Applied interval-transform to upvote_ratio and added transformed upvote_ratio_interval_ to model.\n [-------100%-------] 20000 of 20000 in 1.3 sec. | SPS: 15429.0 | ETA: 0.0Applied interval-transform to upvote_ratio and added transformed upvote_ratio_interval_ to model.\n [-------100%-------] 20000 of 20000 in 1.3 sec. | SPS: 15146.5 | ETA: 0.0"
]
],
[
[
"Some distributions are very tight, others have very long tails (relatively speaking), expressing our uncertainty with what the true upvote ratio might be.\n\n### Sorting!\n\nWe have been ignoring the goal of this exercise: how do we sort the submissions from *best to worst*? Of course, we cannot sort distributions, we must sort scalar numbers. There are many ways to distill a distribution down to a scalar: expressing the distribution through its expected value, or mean, is one way. Choosing the mean is a bad choice though. This is because the mean does not take into account the uncertainty of distributions.\n\nI suggest using the *95% least plausible value*, defined as the value such that there is only a 5% chance the true parameter is lower (think of the lower bound on the 95% credible region). Below are the posterior distributions with the 95% least-plausible value plotted:",
"_____no_output_____"
]
],
[
[
"N = posteriors[0].shape[0]\nlower_limits = []\n\nfor i in range(len(submissions)):\n j = submissions[i]\n plt.hist( posteriors[i], bins = 20, normed = True, alpha = .9, \n histtype=\"step\",color = colours[i], lw = 3,\n label = '(%d up:%d down)\\n%s...'%(votes[j, 0], votes[j,1], contents[j][:50]) )\n plt.hist( posteriors[i], bins = 20, normed = True, alpha = .2, \n histtype=\"stepfilled\",color = colours[i], lw = 3, )\n v = np.sort( posteriors[i] )[ int(0.05*N) ]\n #plt.vlines( v, 0, 15 , color = \"k\", alpha = 1, linewidths=3 )\n plt.vlines( v, 0, 10 , color = colours[i], linestyles = \"--\", linewidths=3 )\n lower_limits.append(v)\n plt.legend(loc=\"upper left\")\n\nplt.legend(loc=\"upper left\")\nplt.title(\"Posterior distributions of upvote ratios on different submissions\");\norder = np.argsort( -np.array( lower_limits ) )\nprint(order, lower_limits)",
"[1 0 2 3] [0.80034320917496615, 0.94092009444598201, 0.74660503350561902, 0.72190353389632911]\n"
]
],
[
[
"The best submissions, according to our procedure, are the submissions that are *most-likely* to score a high percentage of upvotes. Visually those are the submissions with the 95% least plausible value close to 1.\n\nWhy is sorting based on this quantity a good idea? By ordering by the 95% least plausible value, we are being the most conservative with what we think is best. That is, even in the worst case scenario, when we have severely overestimated the upvote ratio, we can be sure the best submissions are still on top. Under this ordering, we impose the following very natural properties:\n\n1. given two submissions with the same observed upvote ratio, we will assign the submission with more votes as better (since we are more confident it has a higher ratio).\n2. given two submissions with the same number of votes, we still assign the submission with more upvotes as *better*.\n\n### But this is too slow for real-time!\n\nI agree, computing the posterior of every submission takes a long time, and by the time you have computed it, likely the data has changed. I delay the mathematics to the appendix, but I suggest using the following formula to compute the lower bound very fast.\n\n$$ \\frac{a}{a + b} - 1.65\\sqrt{ \\frac{ab}{ (a+b)^2(a + b +1 ) } }$$\n\nwhere \n\\begin{align}\n& a = 1 + u \\\\\\\\\n& b = 1 + d \\\\\\\\\n\\end{align}\n\n$u$ is the number of upvotes, and $d$ is the number of downvotes. The formula is a shortcut in Bayesian inference, which will be further explained in Chapter 6 when we discuss priors in more detail.\n",
"_____no_output_____"
]
],
[
[
"def intervals(u,d):\n a = 1. + u\n b = 1. + d\n mu = a/(a+b)\n std_err = 1.65*np.sqrt( (a*b)/( (a+b)**2*(a+b+1.) ) )\n return ( mu, std_err )\n\nprint(\"Approximate lower bounds:\")\nposterior_mean, std_err = intervals(votes[:,0],votes[:,1])\nlb = posterior_mean - std_err\nprint(lb)\nprint(\"\\n\")\nprint(\"Top 40 Sorted according to approximate lower bounds:\")\nprint(\"\\n\")\norder = np.argsort( -lb )\nordered_contents = []\nfor i in order[:40]:\n ordered_contents.append( contents[i] )\n print(votes[i,0], votes[i,1], contents[i])\n print(\"-------------\")",
"Approximate lower bounds:\n[ 0.93349005 0.9532194 0.94149718 0.90859764 0.88705356 0.8558795\n 0.85644927 0.93752679 0.95767101 0.91131012 0.910073 0.915999\n 0.9140058 0.83276025 0.87593961 0.87436674 0.92830849 0.90642832\n 0.89187973 0.89950891 0.91295322 0.78607629 0.90250203 0.79950031\n 0.85219422 0.83703439 0.7619808 0.81301134 0.7313114 0.79137561\n 0.82701445 0.85542404 0.82309334 0.75211374 0.82934814 0.82674958\n 0.80933194 0.87448152 0.85350205 0.75460106 0.82934814 0.74417233\n 0.79924258 0.8189683 0.75460106 0.90744016 0.83838023 0.78802791\n 0.78400654 0.64638659 0.62047936 0.76137738 0.81365241 0.83838023\n 0.78457533 0.84980627 0.79249393 0.69020315 0.69593922 0.70758151\n 0.70268831 0.91620627 0.73346864 0.86382644 0.80877728 0.72708753\n 0.79822085 0.68333632 0.81699014 0.65100453 0.79809005 0.74702492\n 0.77318569 0.83221179 0.66500492 0.68134548 0.7249286 0.59412132\n 0.58191312 0.73142963 0.73142963 0.66251028 0.87152685 0.74107856\n 0.60935684 0.87152685 0.77484517 0.88783675 0.81814153 0.54569789\n 0.6122496 0.75613569 0.53511973 0.74556767 0.81814153 0.85773646\n 0.6122496 0.64814153]\n\n\nTop 40 Sorted according to approximate lower bounds:\n\n\n596 18 Someone should develop an AI specifically for reading Terms & Conditions and flagging dubious parts.\n-------------\n2360 98 Porn is the only industry where it is not only acceptable but standard to separate people based on race, sex and sexual preference.\n-------------\n1918 101 All polls are biased towards people who are willing to take polls\n-------------\n948 50 They should charge less for drinks in the drive-thru because you can't refill them.\n-------------\n3740 239 When I was in elementary school and going through the DARE program, I was positive a gang of older kids was going to corner me and force me to smoke pot. Then I became an adult and realized nobody is giving free drugs to somebody that doesn't want them.\n-------------\n166 7 \"Noted\" is the professional way of saying \"K\".\n-------------\n29 0 Rewatching Mr. Bean, I've realised that the character is an eccentric genius and not a blithering idiot.\n-------------\n289 18 You've been doing weird cameos in your friends' dreams since kindergarten.\n-------------\n269 17 At some point every parent has stopped wiping their child's butt and hoped for the best.\n-------------\n121 6 Is it really fair to say a person over 85 has heart failure? Technically, that heart has done exceptionally well.\n-------------\n535 40 It's surreal to think that the sun and moon and stars we gaze up at are the same objects that have been observed for millenia, by everyone in the history of humanity from cavemen to Aristotle to Jesus to George Washington.\n-------------\n527 40 I wonder if America's internet is censored in a similar way that North Korea's is, but we have no idea of it happening.\n-------------\n1510 131 Kenny's family is poor because they're always paying for his funeral.\n-------------\n43 1 If I was as careful with my whole paycheck as I am with my last $20 I'd be a whole lot better off\n-------------\n162 10 Black hair ties are probably the most popular bracelets in the world.\n-------------\n107 6 The best answer to the interview question \"What is your greatest weakness?\" is \"interviews\".\n-------------\n127 8 Surfing the internet without ads feels like a summer evening without mosquitoes\n-------------\n159 12 I wonder if Superman ever put a pair of glasses on Lois Lane's dog, and she was like \"what's this Clark? Did you get me a new dog?\"\n-------------\n21 0 Sitting on a cold toilet seat or a warm toilet seat both suck for different reasons.\n-------------\n1414 157 My life is really like Rihanna's song, \"just work work work work work\" and the rest of it I can't really understand.\n-------------\n222 22 I'm honestly slightly concerned how often Reddit commenters make me laugh compared to my real life friends.\n-------------\n52 3 The world must have been a spookier place altogether when candles and gas lamps were the only sources of light at night besides the moon and the stars.\n-------------\n194 19 I have not been thankful enough in the last few years that the Black Eyed Peas are no longer ever on the radio\n-------------\n18 0 Living on the coast is having the window seat of the land you live on.\n-------------\n18 0 Binoculars are like walkie talkies for the deaf.\n-------------\n28 1 Now that I am a parent of multiple children I have realized that my parents were lying through their teeth when they said they didn't have a favorite.\n-------------\n16 0 I sneer at people who read tabloids, but every time I look someone up on Wikipedia the first thing I look for is what controversies they've been involved in.\n-------------\n1559 233 Kid's menus at restaurants should be smaller portions of the same adult dishes at lower prices and not the junk food that they usually offer.\n-------------\n1426 213 Eventually once all phones are waterproof we'll be able to push people into pools again\n-------------\n61 5 Myspace is so outdated that jokes about it being outdated has become outdated\n-------------\n52 4 As a kid, seeing someone step on a banana peel and not slip was a disappointment.\n-------------\n90 9 Yahoo!® is the RadioShack® of the Internet.\n-------------\n34 2 People who \"tell it like it is\" rarely do so to say something nice\n-------------\n39 3 Closing your eyes after turning off your alarm is a very dangerous game.\n-------------\n39 3 Your known 'first word' is the first word your parents heard you speak. In reality, it may have been a completely different word you said when you were alone.\n-------------\n87 10 \"Smells Like Teen Spirit\" is as old to listeners of today as \"Yellow Submarine\" was to listeners of 1991.\n-------------\n239 36 if an ocean didnt stop immigrants from coming to America what makes us think a wall will?\n-------------\n22 1 The phonebook was the biggest invasion of privacy that everyone was oddly ok with.\n-------------\n57 6 I'm actually the most productive when I procrastinate because I'm doing everything I possibly can to avoid the main task at hand.\n-------------\n57 6 You will never feel how long time is until you have allergies and snot slowly dripping out of your nostrils, while sitting in a classroom with no tissues.\n-------------\n"
]
],
[
[
"We can view the ordering visually by plotting the posterior mean and bounds, and sorting by the lower bound. In the plot below, notice that the left error-bar is sorted (as we suggested this is the best way to determine an ordering), so the means, indicated by dots, do not follow any strong pattern. ",
"_____no_output_____"
]
],
[
[
"r_order = order[::-1][-40:]\nplt.errorbar( posterior_mean[r_order], np.arange( len(r_order) ), \n xerr=std_err[r_order], capsize=0, fmt=\"o\",\n color = \"#7A68A6\")\nplt.xlim( 0.3, 1)\nplt.yticks( np.arange( len(r_order)-1,-1,-1 ), map( lambda x: x[:30].replace(\"\\n\",\"\"), ordered_contents) );",
"_____no_output_____"
]
],
[
[
"In the graphic above, you can see why sorting by mean would be sub-optimal.",
"_____no_output_____"
],
[
"### Extension to Starred rating systems\n\nThe above procedure works well for upvote-downvotes schemes, but what about systems that use star ratings, e.g. 5 star rating systems. Similar problems apply with simply taking the average: an item with two perfect ratings would beat an item with thousands of perfect ratings, but a single sub-perfect rating. \n\n\nWe can consider the upvote-downvote problem above as binary: 0 is a downvote, 1 if an upvote. A $N$-star rating system can be seen as a more continuous version of above, and we can set $n$ stars rewarded is equivalent to rewarding $\\frac{n}{N}$. For example, in a 5-star system, a 2 star rating corresponds to 0.4. A perfect rating is a 1. We can use the same formula as before, but with $a,b$ defined differently:\n\n\n$$ \\frac{a}{a + b} - 1.65\\sqrt{ \\frac{ab}{ (a+b)^2(a + b +1 ) } }$$\n\nwhere \n\n\\begin{align}\n& a = 1 + S \\\\\\\\\n& b = 1 + N - S \\\\\\\\\n\\end{align}\n\nwhere $N$ is the number of users who rated, and $S$ is the sum of all the ratings, under the equivalence scheme mentioned above. ",
"_____no_output_____"
],
[
"##### Example: Counting Github stars\n\nWhat is the average number of stars a Github repository has? How would you calculate this? There are over 6 million respositories, so there is more than enough data to invoke the Law of Large numbers. Let's start pulling some data. TODO",
"_____no_output_____"
],
[
"### Conclusion\n\nWhile the Law of Large Numbers is cool, it is only true so much as its name implies: with large sample sizes only. We have seen how our inference can be affected by not considering *how the data is shaped*. \n\n1. By (cheaply) drawing many samples from the posterior distributions, we can ensure that the Law of Large Number applies as we approximate expected values (which we will do in the next chapter).\n\n2. Bayesian inference understands that with small sample sizes, we can observe wild randomness. Our posterior distribution will reflect this by being more spread rather than tightly concentrated. Thus, our inference should be correctable.\n\n3. There are major implications of not considering the sample size, and trying to sort objects that are unstable leads to pathological orderings. The method provided above solves this problem.\n",
"_____no_output_____"
],
[
"### Appendix\n\n##### Derivation of sorting submissions formula\n\nBasically what we are doing is using a Beta prior (with parameters $a=1, b=1$, which is a uniform distribution), and using a Binomial likelihood with observations $u, N = u+d$. This means our posterior is a Beta distribution with parameters $a' = 1 + u, b' = 1 + (N - u) = 1+d$. We then need to find the value, $x$, such that 0.05 probability is less than $x$. This is usually done by inverting the CDF ([Cumulative Distribution Function](http://en.wikipedia.org/wiki/Cumulative_Distribution_Function)), but the CDF of the beta, for integer parameters, is known but is a large sum [3]. \n\nWe instead use a Normal approximation. The mean of the Beta is $\\mu = a'/(a'+b')$ and the variance is \n\n$$\\sigma^2 = \\frac{a'b'}{ (a' + b')^2(a'+b'+1) }$$\n\nHence we solve the following equation for $x$ and have an approximate lower bound. \n\n$$ 0.05 = \\Phi\\left( \\frac{(x - \\mu)}{\\sigma}\\right) $$ \n\n$\\Phi$ being the [cumulative distribution for the normal distribution](http://en.wikipedia.org/wiki/Normal_distribution#Cumulative_distribution)\n\n\n\n\n",
"_____no_output_____"
],
[
"##### Exercises\n\n1\\. How would you estimate the quantity $E\\left[ \\cos{X} \\right]$, where $X \\sim \\text{Exp}(4)$? What about $E\\left[ \\cos{X} | X \\lt 1\\right]$, i.e. the expected value *given* we know $X$ is less than 1? Would you need more samples than the original samples size to be equally accurate?",
"_____no_output_____"
]
],
[
[
"## Enter code here\nimport scipy.stats as stats\nexp = stats.expon( scale=4 )\nN = 1e5\nX = exp.rvs( int(N) )\n## ...",
"_____no_output_____"
]
],
[
[
"2\\. The following table was located in the paper \"Going for Three: Predicting the Likelihood of Field Goal Success with Logistic Regression\" [2]. The table ranks football field-goal kickers by their percent of non-misses. What mistake have the researchers made?\n\n-----\n\n#### Kicker Careers Ranked by Make Percentage\n<table><tbody><tr><th>Rank </th><th>Kicker </th><th>Make % </th><th>Number of Kicks</th></tr><tr><td>1 </td><td>Garrett Hartley </td><td>87.7 </td><td>57</td></tr><tr><td>2</td><td> Matt Stover </td><td>86.8 </td><td>335</td></tr><tr><td>3 </td><td>Robbie Gould </td><td>86.2 </td><td>224</td></tr><tr><td>4 </td><td>Rob Bironas </td><td>86.1 </td><td>223</td></tr><tr><td>5</td><td> Shayne Graham </td><td>85.4 </td><td>254</td></tr><tr><td>… </td><td>… </td><td>…</td><td> </td></tr><tr><td>51</td><td> Dave Rayner </td><td>72.2 </td><td>90</td></tr><tr><td>52</td><td> Nick Novak </td><td>71.9 </td><td>64</td></tr><tr><td>53 </td><td>Tim Seder </td><td>71.0 </td><td>62</td></tr><tr><td>54 </td><td>Jose Cortez </td><td>70.7</td><td> 75</td></tr><tr><td>55 </td><td>Wade Richey </td><td>66.1</td><td> 56</td></tr></tbody></table>",
"_____no_output_____"
],
[
"In August 2013, [a popular post](http://bpodgursky.wordpress.com/2013/08/21/average-income-per-programming-language/) on the average income per programmer of different languages was trending. Here's the summary chart: (reproduced without permission, cause when you lie with stats, you gunna get the hammer). What do you notice about the extremes?\n\n------\n\n#### Average household income by programming language\n\n<table >\n <tr><td>Language</td><td>Average Household Income ($)</td><td>Data Points</td></tr>\n <tr><td>Puppet</td><td>87,589.29</td><td>112</td></tr>\n <tr><td>Haskell</td><td>89,973.82</td><td>191</td></tr>\n <tr><td>PHP</td><td>94,031.19</td><td>978</td></tr>\n <tr><td>CoffeeScript</td><td>94,890.80</td><td>435</td></tr>\n <tr><td>VimL</td><td>94,967.11</td><td>532</td></tr>\n <tr><td>Shell</td><td>96,930.54</td><td>979</td></tr>\n <tr><td>Lua</td><td>96,930.69</td><td>101</td></tr>\n <tr><td>Erlang</td><td>97,306.55</td><td>168</td></tr>\n <tr><td>Clojure</td><td>97,500.00</td><td>269</td></tr>\n <tr><td>Python</td><td>97,578.87</td><td>2314</td></tr>\n <tr><td>JavaScript</td><td>97,598.75</td><td>3443</td></tr>\n <tr><td>Emacs Lisp</td><td>97,774.65</td><td>355</td></tr>\n <tr><td>C#</td><td>97,823.31</td><td>665</td></tr>\n <tr><td>Ruby</td><td>98,238.74</td><td>3242</td></tr>\n <tr><td>C++</td><td>99,147.93</td><td>845</td></tr>\n <tr><td>CSS</td><td>99,881.40</td><td>527</td></tr>\n <tr><td>Perl</td><td>100,295.45</td><td>990</td></tr>\n <tr><td>C</td><td>100,766.51</td><td>2120</td></tr>\n <tr><td>Go</td><td>101,158.01</td><td>231</td></tr>\n <tr><td>Scala</td><td>101,460.91</td><td>243</td></tr>\n <tr><td>ColdFusion</td><td>101,536.70</td><td>109</td></tr>\n <tr><td>Objective-C</td><td>101,801.60</td><td>562</td></tr>\n <tr><td>Groovy</td><td>102,650.86</td><td>116</td></tr>\n <tr><td>Java</td><td>103,179.39</td><td>1402</td></tr>\n <tr><td>XSLT</td><td>106,199.19</td><td>123</td></tr>\n <tr><td>ActionScript</td><td>108,119.47</td><td>113</td></tr>\n</table>",
"_____no_output_____"
],
[
"### References\n\n1. Wainer, Howard. *The Most Dangerous Equation*. American Scientist, Volume 95.\n2. Clarck, Torin K., Aaron W. Johnson, and Alexander J. Stimpson. \"Going for Three: Predicting the Likelihood of Field Goal Success with Logistic Regression.\" (2013): n. page. [Web](http://www.sloansportsconference.com/wp-content/uploads/2013/Going%20for%20Three%20Predicting%20the%20Likelihood%20of%20Field%20Goal%20Success%20with%20Logistic%20Regression.pdf). 20 Feb. 2013.\n3. http://en.wikipedia.org/wiki/Beta_function#Incomplete_beta_function",
"_____no_output_____"
]
],
[
[
"from IPython.core.display import HTML\ndef css_styling():\n styles = open(\"../styles/custom.css\", \"r\").read()\n return HTML(styles)\ncss_styling()",
"_____no_output_____"
]
],
[
[
"<style>\n img{\n max-width:800px}\n</style>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d002f5490e14573b2c804c59f9f79321382025c6 | 281,255 | ipynb | Jupyter Notebook | experiments/baseline_ptn/wisig/trials/2/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | experiments/baseline_ptn/wisig/trials/2/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | experiments/baseline_ptn/wisig/trials/2/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | 75.524973 | 76,916 | 0.731838 | [
[
[
"# PTN Template\nThis notebook serves as a template for single dataset PTN experiments \nIt can be run on its own by setting STANDALONE to True (do a find for \"STANDALONE\" to see where) \nBut it is intended to be executed as part of a *papermill.py script. See any of the \nexperimentes with a papermill script to get started with that workflow. ",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n%matplotlib inline\n\n \nimport os, json, sys, time, random\nimport numpy as np\nimport torch\nfrom torch.optim import Adam\nfrom easydict import EasyDict\nimport matplotlib.pyplot as plt\n\nfrom steves_models.steves_ptn import Steves_Prototypical_Network\n\nfrom steves_utils.lazy_iterable_wrapper import Lazy_Iterable_Wrapper\nfrom steves_utils.iterable_aggregator import Iterable_Aggregator\nfrom steves_utils.ptn_train_eval_test_jig import PTN_Train_Eval_Test_Jig\nfrom steves_utils.torch_sequential_builder import build_sequential\nfrom steves_utils.torch_utils import get_dataset_metrics, ptn_confusion_by_domain_over_dataloader\nfrom steves_utils.utils_v2 import (per_domain_accuracy_from_confusion, get_datasets_base_path)\nfrom steves_utils.PTN.utils import independent_accuracy_assesment\n\nfrom steves_utils.stratified_dataset.episodic_accessor import Episodic_Accessor_Factory\n\nfrom steves_utils.ptn_do_report import (\n get_loss_curve,\n get_results_table,\n get_parameters_table,\n get_domain_accuracies,\n)\n\nfrom steves_utils.transforms import get_chained_transform",
"_____no_output_____"
]
],
[
[
"# Required Parameters\nThese are allowed parameters, not defaults\nEach of these values need to be present in the injected parameters (the notebook will raise an exception if they are not present)\n\nPapermill uses the cell tag \"parameters\" to inject the real parameters below this cell.\nEnable tags to see what I mean",
"_____no_output_____"
]
],
[
[
"required_parameters = {\n \"experiment_name\",\n \"lr\",\n \"device\",\n \"seed\",\n \"dataset_seed\",\n \"labels_source\",\n \"labels_target\",\n \"domains_source\",\n \"domains_target\",\n \"num_examples_per_domain_per_label_source\",\n \"num_examples_per_domain_per_label_target\",\n \"n_shot\",\n \"n_way\",\n \"n_query\",\n \"train_k_factor\",\n \"val_k_factor\",\n \"test_k_factor\",\n \"n_epoch\",\n \"patience\",\n \"criteria_for_best\",\n \"x_transforms_source\",\n \"x_transforms_target\",\n \"episode_transforms_source\",\n \"episode_transforms_target\",\n \"pickle_name\",\n \"x_net\",\n \"NUM_LOGS_PER_EPOCH\",\n \"BEST_MODEL_PATH\",\n \"torch_default_dtype\"\n}",
"_____no_output_____"
],
[
"\n\nstandalone_parameters = {}\nstandalone_parameters[\"experiment_name\"] = \"STANDALONE PTN\"\nstandalone_parameters[\"lr\"] = 0.0001\nstandalone_parameters[\"device\"] = \"cuda\"\n\nstandalone_parameters[\"seed\"] = 1337\nstandalone_parameters[\"dataset_seed\"] = 1337\n\n\nstandalone_parameters[\"num_examples_per_domain_per_label_source\"]=100\nstandalone_parameters[\"num_examples_per_domain_per_label_target\"]=100\n\nstandalone_parameters[\"n_shot\"] = 3\nstandalone_parameters[\"n_query\"] = 2\nstandalone_parameters[\"train_k_factor\"] = 1\nstandalone_parameters[\"val_k_factor\"] = 2\nstandalone_parameters[\"test_k_factor\"] = 2\n\n\nstandalone_parameters[\"n_epoch\"] = 100\n\nstandalone_parameters[\"patience\"] = 10\nstandalone_parameters[\"criteria_for_best\"] = \"target_accuracy\"\n\nstandalone_parameters[\"x_transforms_source\"] = [\"unit_power\"]\nstandalone_parameters[\"x_transforms_target\"] = [\"unit_power\"]\nstandalone_parameters[\"episode_transforms_source\"] = []\nstandalone_parameters[\"episode_transforms_target\"] = []\n\nstandalone_parameters[\"torch_default_dtype\"] = \"torch.float32\" \n\n\n\nstandalone_parameters[\"x_net\"] = [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\":[-1, 1, 2, 256]}},\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":1, \"out_channels\":256, \"kernel_size\":(1,7), \"bias\":False, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":256, \"out_channels\":80, \"kernel_size\":(2,7), \"bias\":True, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 80*256, \"out_features\": 256}}, # 80 units per IQ pair\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n]\n\n# Parameters relevant to results\n# These parameters will basically never need to change\nstandalone_parameters[\"NUM_LOGS_PER_EPOCH\"] = 10\nstandalone_parameters[\"BEST_MODEL_PATH\"] = \"./best_model.pth\"\n\n# uncomment for CORES dataset\nfrom steves_utils.CORES.utils import (\n ALL_NODES,\n ALL_NODES_MINIMUM_1000_EXAMPLES,\n ALL_DAYS\n)\n\n\nstandalone_parameters[\"labels_source\"] = ALL_NODES\nstandalone_parameters[\"labels_target\"] = ALL_NODES\n\nstandalone_parameters[\"domains_source\"] = [1]\nstandalone_parameters[\"domains_target\"] = [2,3,4,5]\n\nstandalone_parameters[\"pickle_name\"] = \"cores.stratified_ds.2022A.pkl\"\n\n\n# Uncomment these for ORACLE dataset\n# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n# standalone_parameters[\"labels_source\"] = ALL_SERIAL_NUMBERS\n# standalone_parameters[\"labels_target\"] = ALL_SERIAL_NUMBERS\n# standalone_parameters[\"domains_source\"] = [8,20, 38,50]\n# standalone_parameters[\"domains_target\"] = [14, 26, 32, 44, 56]\n# standalone_parameters[\"pickle_name\"] = \"oracle.frame_indexed.stratified_ds.2022A.pkl\"\n# standalone_parameters[\"num_examples_per_domain_per_label_source\"]=1000\n# standalone_parameters[\"num_examples_per_domain_per_label_target\"]=1000\n\n# Uncomment these for Metahan dataset\n# standalone_parameters[\"labels_source\"] = list(range(19))\n# standalone_parameters[\"labels_target\"] = list(range(19))\n# standalone_parameters[\"domains_source\"] = [0]\n# standalone_parameters[\"domains_target\"] = [1]\n# standalone_parameters[\"pickle_name\"] = \"metehan.stratified_ds.2022A.pkl\"\n# standalone_parameters[\"n_way\"] = len(standalone_parameters[\"labels_source\"])\n# standalone_parameters[\"num_examples_per_domain_per_label_source\"]=200\n# standalone_parameters[\"num_examples_per_domain_per_label_target\"]=100\n\n\nstandalone_parameters[\"n_way\"] = len(standalone_parameters[\"labels_source\"])",
"_____no_output_____"
],
[
"# Parameters\nparameters = {\n \"experiment_name\": \"baseline_ptn_wisig\",\n \"lr\": 0.001,\n \"device\": \"cuda\",\n \"seed\": 1337,\n \"dataset_seed\": 1337,\n \"labels_source\": [\n \"1-10\",\n \"1-12\",\n \"1-14\",\n \"1-16\",\n \"1-18\",\n \"1-19\",\n \"1-8\",\n \"10-11\",\n \"10-17\",\n \"10-4\",\n \"10-7\",\n \"11-1\",\n \"11-10\",\n \"11-19\",\n \"11-20\",\n \"11-4\",\n \"11-7\",\n \"12-19\",\n \"12-20\",\n \"12-7\",\n \"13-14\",\n \"13-18\",\n \"13-19\",\n \"13-20\",\n \"13-3\",\n \"13-7\",\n \"14-10\",\n \"14-11\",\n \"14-12\",\n \"14-13\",\n \"14-14\",\n \"14-19\",\n \"14-20\",\n \"14-7\",\n \"14-8\",\n \"14-9\",\n \"15-1\",\n \"15-19\",\n \"15-6\",\n \"16-1\",\n \"16-16\",\n \"16-19\",\n \"16-20\",\n \"17-10\",\n \"17-11\",\n \"18-1\",\n \"18-10\",\n \"18-11\",\n \"18-12\",\n \"18-13\",\n \"18-14\",\n \"18-15\",\n \"18-16\",\n \"18-17\",\n \"18-19\",\n \"18-2\",\n \"18-20\",\n \"18-4\",\n \"18-5\",\n \"18-7\",\n \"18-8\",\n \"18-9\",\n \"19-1\",\n \"19-10\",\n \"19-11\",\n \"19-12\",\n \"19-13\",\n \"19-14\",\n \"19-15\",\n \"19-19\",\n \"19-2\",\n \"19-20\",\n \"19-3\",\n \"19-4\",\n \"19-6\",\n \"19-7\",\n \"19-8\",\n \"19-9\",\n \"2-1\",\n \"2-13\",\n \"2-15\",\n \"2-3\",\n \"2-4\",\n \"2-5\",\n \"2-6\",\n \"2-7\",\n \"2-8\",\n \"20-1\",\n \"20-12\",\n \"20-14\",\n \"20-15\",\n \"20-16\",\n \"20-18\",\n \"20-19\",\n \"20-20\",\n \"20-3\",\n \"20-4\",\n \"20-5\",\n \"20-7\",\n \"20-8\",\n \"3-1\",\n \"3-13\",\n \"3-18\",\n \"3-2\",\n \"3-8\",\n \"4-1\",\n \"4-10\",\n \"4-11\",\n \"5-1\",\n \"5-5\",\n \"6-1\",\n \"6-15\",\n \"6-6\",\n \"7-10\",\n \"7-11\",\n \"7-12\",\n \"7-13\",\n \"7-14\",\n \"7-7\",\n \"7-8\",\n \"7-9\",\n \"8-1\",\n \"8-13\",\n \"8-14\",\n \"8-18\",\n \"8-20\",\n \"8-3\",\n \"8-8\",\n \"9-1\",\n \"9-7\",\n ],\n \"labels_target\": [\n \"1-10\",\n \"1-12\",\n \"1-14\",\n \"1-16\",\n \"1-18\",\n \"1-19\",\n \"1-8\",\n \"10-11\",\n \"10-17\",\n \"10-4\",\n \"10-7\",\n \"11-1\",\n \"11-10\",\n \"11-19\",\n \"11-20\",\n \"11-4\",\n \"11-7\",\n \"12-19\",\n \"12-20\",\n \"12-7\",\n \"13-14\",\n \"13-18\",\n \"13-19\",\n \"13-20\",\n \"13-3\",\n \"13-7\",\n \"14-10\",\n \"14-11\",\n \"14-12\",\n \"14-13\",\n \"14-14\",\n \"14-19\",\n \"14-20\",\n \"14-7\",\n \"14-8\",\n \"14-9\",\n \"15-1\",\n \"15-19\",\n \"15-6\",\n \"16-1\",\n \"16-16\",\n \"16-19\",\n \"16-20\",\n \"17-10\",\n \"17-11\",\n \"18-1\",\n \"18-10\",\n \"18-11\",\n \"18-12\",\n \"18-13\",\n \"18-14\",\n \"18-15\",\n \"18-16\",\n \"18-17\",\n \"18-19\",\n \"18-2\",\n \"18-20\",\n \"18-4\",\n \"18-5\",\n \"18-7\",\n \"18-8\",\n \"18-9\",\n \"19-1\",\n \"19-10\",\n \"19-11\",\n \"19-12\",\n \"19-13\",\n \"19-14\",\n \"19-15\",\n \"19-19\",\n \"19-2\",\n \"19-20\",\n \"19-3\",\n \"19-4\",\n \"19-6\",\n \"19-7\",\n \"19-8\",\n \"19-9\",\n \"2-1\",\n \"2-13\",\n \"2-15\",\n \"2-3\",\n \"2-4\",\n \"2-5\",\n \"2-6\",\n \"2-7\",\n \"2-8\",\n \"20-1\",\n \"20-12\",\n \"20-14\",\n \"20-15\",\n \"20-16\",\n \"20-18\",\n \"20-19\",\n \"20-20\",\n \"20-3\",\n \"20-4\",\n \"20-5\",\n \"20-7\",\n \"20-8\",\n \"3-1\",\n \"3-13\",\n \"3-18\",\n \"3-2\",\n \"3-8\",\n \"4-1\",\n \"4-10\",\n \"4-11\",\n \"5-1\",\n \"5-5\",\n \"6-1\",\n \"6-15\",\n \"6-6\",\n \"7-10\",\n \"7-11\",\n \"7-12\",\n \"7-13\",\n \"7-14\",\n \"7-7\",\n \"7-8\",\n \"7-9\",\n \"8-1\",\n \"8-13\",\n \"8-14\",\n \"8-18\",\n \"8-20\",\n \"8-3\",\n \"8-8\",\n \"9-1\",\n \"9-7\",\n ],\n \"x_transforms_source\": [],\n \"x_transforms_target\": [],\n \"episode_transforms_source\": [],\n \"episode_transforms_target\": [],\n \"num_examples_per_domain_per_label_source\": 100,\n \"num_examples_per_domain_per_label_target\": 100,\n \"n_shot\": 3,\n \"n_way\": 130,\n \"n_query\": 2,\n \"train_k_factor\": 1,\n \"val_k_factor\": 2,\n \"test_k_factor\": 2,\n \"torch_default_dtype\": \"torch.float64\",\n \"n_epoch\": 50,\n \"patience\": 3,\n \"criteria_for_best\": \"target_loss\",\n \"x_net\": [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\": [-1, 1, 2, 256]}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 1,\n \"out_channels\": 256,\n \"kernel_size\": [1, 7],\n \"bias\": False,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 256}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 256,\n \"out_channels\": 80,\n \"kernel_size\": [2, 7],\n \"bias\": True,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 20480, \"out_features\": 256}},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\": 256}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n ],\n \"NUM_LOGS_PER_EPOCH\": 10,\n \"BEST_MODEL_PATH\": \"./best_model.pth\",\n \"pickle_name\": \"wisig.node3-19.stratified_ds.2022A.pkl\",\n \"domains_source\": [3],\n \"domains_target\": [1, 2, 4],\n}\n",
"_____no_output_____"
],
[
"# Set this to True if you want to run this template directly\nSTANDALONE = False\nif STANDALONE:\n print(\"parameters not injected, running with standalone_parameters\")\n parameters = standalone_parameters\n\nif not 'parameters' in locals() and not 'parameters' in globals():\n raise Exception(\"Parameter injection failed\")\n\n#Use an easy dict for all the parameters\np = EasyDict(parameters)\n\nsupplied_keys = set(p.keys())\n\nif supplied_keys != required_parameters:\n print(\"Parameters are incorrect\")\n if len(supplied_keys - required_parameters)>0: print(\"Shouldn't have:\", str(supplied_keys - required_parameters))\n if len(required_parameters - supplied_keys)>0: print(\"Need to have:\", str(required_parameters - supplied_keys))\n raise RuntimeError(\"Parameters are incorrect\")\n\n",
"_____no_output_____"
],
[
"###################################\n# Set the RNGs and make it all deterministic\n###################################\nnp.random.seed(p.seed)\nrandom.seed(p.seed)\ntorch.manual_seed(p.seed)\n\ntorch.use_deterministic_algorithms(True) ",
"_____no_output_____"
],
[
"###########################################\n# The stratified datasets honor this\n###########################################\ntorch.set_default_dtype(eval(p.torch_default_dtype))",
"_____no_output_____"
],
[
"###################################\n# Build the network(s)\n# Note: It's critical to do this AFTER setting the RNG\n# (This is due to the randomized initial weights)\n###################################\nx_net = build_sequential(p.x_net)",
"_____no_output_____"
],
[
"start_time_secs = time.time()",
"_____no_output_____"
],
[
"###################################\n# Build the dataset\n###################################\n\nif p.x_transforms_source == []: x_transform_source = None\nelse: x_transform_source = get_chained_transform(p.x_transforms_source) \n\nif p.x_transforms_target == []: x_transform_target = None\nelse: x_transform_target = get_chained_transform(p.x_transforms_target)\n\nif p.episode_transforms_source == []: episode_transform_source = None\nelse: raise Exception(\"episode_transform_source not implemented\")\n\nif p.episode_transforms_target == []: episode_transform_target = None\nelse: raise Exception(\"episode_transform_target not implemented\")\n\n\neaf_source = Episodic_Accessor_Factory(\n labels=p.labels_source,\n domains=p.domains_source,\n num_examples_per_domain_per_label=p.num_examples_per_domain_per_label_source,\n iterator_seed=p.seed,\n dataset_seed=p.dataset_seed,\n n_shot=p.n_shot,\n n_way=p.n_way,\n n_query=p.n_query,\n train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),\n pickle_path=os.path.join(get_datasets_base_path(), p.pickle_name),\n x_transform_func=x_transform_source,\n example_transform_func=episode_transform_source,\n \n)\ntrain_original_source, val_original_source, test_original_source = eaf_source.get_train(), eaf_source.get_val(), eaf_source.get_test()\n\n\neaf_target = Episodic_Accessor_Factory(\n labels=p.labels_target,\n domains=p.domains_target,\n num_examples_per_domain_per_label=p.num_examples_per_domain_per_label_target,\n iterator_seed=p.seed,\n dataset_seed=p.dataset_seed,\n n_shot=p.n_shot,\n n_way=p.n_way,\n n_query=p.n_query,\n train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),\n pickle_path=os.path.join(get_datasets_base_path(), p.pickle_name),\n x_transform_func=x_transform_target,\n example_transform_func=episode_transform_target,\n)\ntrain_original_target, val_original_target, test_original_target = eaf_target.get_train(), eaf_target.get_val(), eaf_target.get_test()\n\n\ntransform_lambda = lambda ex: ex[1] # Original is (<domain>, <episode>) so we strip down to episode only\n\ntrain_processed_source = Lazy_Iterable_Wrapper(train_original_source, transform_lambda)\nval_processed_source = Lazy_Iterable_Wrapper(val_original_source, transform_lambda)\ntest_processed_source = Lazy_Iterable_Wrapper(test_original_source, transform_lambda)\n\ntrain_processed_target = Lazy_Iterable_Wrapper(train_original_target, transform_lambda)\nval_processed_target = Lazy_Iterable_Wrapper(val_original_target, transform_lambda)\ntest_processed_target = Lazy_Iterable_Wrapper(test_original_target, transform_lambda)\n\ndatasets = EasyDict({\n \"source\": {\n \"original\": {\"train\":train_original_source, \"val\":val_original_source, \"test\":test_original_source},\n \"processed\": {\"train\":train_processed_source, \"val\":val_processed_source, \"test\":test_processed_source}\n },\n \"target\": {\n \"original\": {\"train\":train_original_target, \"val\":val_original_target, \"test\":test_original_target},\n \"processed\": {\"train\":train_processed_target, \"val\":val_processed_target, \"test\":test_processed_target}\n },\n})",
"_____no_output_____"
],
[
"# Some quick unit tests on the data\nfrom steves_utils.transforms import get_average_power, get_average_magnitude\n\nq_x, q_y, s_x, s_y, truth = next(iter(train_processed_source))\n\nassert q_x.dtype == eval(p.torch_default_dtype)\nassert s_x.dtype == eval(p.torch_default_dtype)\n\nprint(\"Visually inspect these to see if they line up with expected values given the transforms\")\nprint('x_transforms_source', p.x_transforms_source)\nprint('x_transforms_target', p.x_transforms_target)\nprint(\"Average magnitude, source:\", get_average_magnitude(q_x[0].numpy()))\nprint(\"Average power, source:\", get_average_power(q_x[0].numpy()))\n\nq_x, q_y, s_x, s_y, truth = next(iter(train_processed_target))\nprint(\"Average magnitude, target:\", get_average_magnitude(q_x[0].numpy()))\nprint(\"Average power, target:\", get_average_power(q_x[0].numpy()))\n",
"Visually inspect these to see if they line up with expected values given the transforms\nx_transforms_source []\nx_transforms_target []\nAverage magnitude, source: 0.07199795071703695\nAverage power, source: 0.006306497542264809\n"
],
[
"###################################\n# Build the model\n###################################\nmodel = Steves_Prototypical_Network(x_net, device=p.device, x_shape=(2,256))\noptimizer = Adam(params=model.parameters(), lr=p.lr)",
"(2, 256)\n"
],
[
"###################################\n# train\n###################################\njig = PTN_Train_Eval_Test_Jig(model, p.BEST_MODEL_PATH, p.device)\n\njig.train(\n train_iterable=datasets.source.processed.train,\n source_val_iterable=datasets.source.processed.val,\n target_val_iterable=datasets.target.processed.val,\n num_epochs=p.n_epoch,\n num_logs_per_epoch=p.NUM_LOGS_PER_EPOCH,\n patience=p.patience,\n optimizer=optimizer,\n criteria_for_best=p.criteria_for_best,\n)",
"epoch: 1, [batch: 1 / 14], examples_per_second: 284.0435, train_label_loss: 3.5532, \n"
],
[
"total_experiment_time_secs = time.time() - start_time_secs",
"_____no_output_____"
],
[
"###################################\n# Evaluate the model\n###################################\nsource_test_label_accuracy, source_test_label_loss = jig.test(datasets.source.processed.test)\ntarget_test_label_accuracy, target_test_label_loss = jig.test(datasets.target.processed.test)\n\nsource_val_label_accuracy, source_val_label_loss = jig.test(datasets.source.processed.val)\ntarget_val_label_accuracy, target_val_label_loss = jig.test(datasets.target.processed.val)\n\nhistory = jig.get_history()\n\ntotal_epochs_trained = len(history[\"epoch_indices\"])\n\nval_dl = Iterable_Aggregator((datasets.source.original.val,datasets.target.original.val))\n\nconfusion = ptn_confusion_by_domain_over_dataloader(model, p.device, val_dl)\nper_domain_accuracy = per_domain_accuracy_from_confusion(confusion)\n\n# Add a key to per_domain_accuracy for if it was a source domain\nfor domain, accuracy in per_domain_accuracy.items():\n per_domain_accuracy[domain] = {\n \"accuracy\": accuracy,\n \"source?\": domain in p.domains_source\n }\n\n# Do an independent accuracy assesment JUST TO BE SURE!\n# _source_test_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.test, p.device)\n# _target_test_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.test, p.device)\n# _source_val_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.val, p.device)\n# _target_val_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.val, p.device)\n\n# assert(_source_test_label_accuracy == source_test_label_accuracy)\n# assert(_target_test_label_accuracy == target_test_label_accuracy)\n# assert(_source_val_label_accuracy == source_val_label_accuracy)\n# assert(_target_val_label_accuracy == target_val_label_accuracy)\n\nexperiment = {\n \"experiment_name\": p.experiment_name,\n \"parameters\": dict(p),\n \"results\": {\n \"source_test_label_accuracy\": source_test_label_accuracy,\n \"source_test_label_loss\": source_test_label_loss,\n \"target_test_label_accuracy\": target_test_label_accuracy,\n \"target_test_label_loss\": target_test_label_loss,\n \"source_val_label_accuracy\": source_val_label_accuracy,\n \"source_val_label_loss\": source_val_label_loss,\n \"target_val_label_accuracy\": target_val_label_accuracy,\n \"target_val_label_loss\": target_val_label_loss,\n \"total_epochs_trained\": total_epochs_trained,\n \"total_experiment_time_secs\": total_experiment_time_secs,\n \"confusion\": confusion,\n \"per_domain_accuracy\": per_domain_accuracy,\n },\n \"history\": history,\n \"dataset_metrics\": get_dataset_metrics(datasets, \"ptn\"),\n}",
"_____no_output_____"
],
[
"ax = get_loss_curve(experiment)\nplt.show()",
"_____no_output_____"
],
[
"get_results_table(experiment)",
"_____no_output_____"
],
[
"get_domain_accuracies(experiment)",
"_____no_output_____"
],
[
"print(\"Source Test Label Accuracy:\", experiment[\"results\"][\"source_test_label_accuracy\"], \"Target Test Label Accuracy:\", experiment[\"results\"][\"target_test_label_accuracy\"])\nprint(\"Source Val Label Accuracy:\", experiment[\"results\"][\"source_val_label_accuracy\"], \"Target Val Label Accuracy:\", experiment[\"results\"][\"target_val_label_accuracy\"])",
"Source Test Label Accuracy: 0.9141025641025641 Target Test Label Accuracy: 0.8427350427350427\nSource Val Label Accuracy: 0.9108974358974359 Target Val Label Accuracy: 0.847008547008547\n"
],
[
"json.dumps(experiment)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d003075cd5ca7d2206d34b0a876c67c7596b94e6 | 11,024 | ipynb | Jupyter Notebook | source/pytorch/deepLearningIn60mins/neural_networks_tutorial.ipynb | alphajayGithub/ai.online | 3e440d88111627827456aa8672516eb389a68e98 | [
"MIT"
] | null | null | null | source/pytorch/deepLearningIn60mins/neural_networks_tutorial.ipynb | alphajayGithub/ai.online | 3e440d88111627827456aa8672516eb389a68e98 | [
"MIT"
] | null | null | null | source/pytorch/deepLearningIn60mins/neural_networks_tutorial.ipynb | alphajayGithub/ai.online | 3e440d88111627827456aa8672516eb389a68e98 | [
"MIT"
] | null | null | null | 58.951872 | 1,918 | 0.613389 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\nNeural Networks\n===============\n\nNeural networks can be constructed using the ``torch.nn`` package.\n\nNow that you had a glimpse of ``autograd``, ``nn`` depends on\n``autograd`` to define models and differentiate them.\nAn ``nn.Module`` contains layers, and a method ``forward(input)`` that\nreturns the ``output``.\n\nFor example, look at this network that classifies digit images:\n\n.. figure:: /_static/img/mnist.png\n :alt: convnet\n\n convnet\n\nIt is a simple feed-forward network. It takes the input, feeds it\nthrough several layers one after the other, and then finally gives the\noutput.\n\nA typical training procedure for a neural network is as follows:\n\n- Define the neural network that has some learnable parameters (or\n weights)\n- Iterate over a dataset of inputs\n- Process input through the network\n- Compute the loss (how far is the output from being correct)\n- Propagate gradients back into the network’s parameters\n- Update the weights of the network, typically using a simple update rule:\n ``weight = weight - learning_rate * gradient``\n\nDefine the network\n------------------\n\nLet’s define this network:\n\n",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\n\n\nclass Net(nn.Module):\n\n def __init__(self):\n super(Net, self).__init__()\n # 1 input image channel, 6 output channels, 5x5 square convolution\n # kernel\n self.conv1 = nn.Conv2d(1, 6, 5)\n self.conv2 = nn.Conv2d(6, 16, 5)\n # an affine operation: y = Wx + b\n self.fc1 = nn.Linear(16 * 5 * 5, 120) # 5*5 from image dimension \n self.fc2 = nn.Linear(120, 84)\n self.fc3 = nn.Linear(84, 10)\n\n def forward(self, x):\n # Max pooling over a (2, 2) window\n x = F.max_pool2d(F.relu(self.conv1(x)), (2, 2))\n # If the size is a square, you can specify with a single number\n x = F.max_pool2d(F.relu(self.conv2(x)), 2)\n x = torch.flatten(x, 1) # flatten all dimensions except the batch dimension\n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n x = self.fc3(x)\n return x\n\n\nnet = Net()\nprint(net)",
"_____no_output_____"
]
],
[
[
"You just have to define the ``forward`` function, and the ``backward``\nfunction (where gradients are computed) is automatically defined for you\nusing ``autograd``.\nYou can use any of the Tensor operations in the ``forward`` function.\n\nThe learnable parameters of a model are returned by ``net.parameters()``\n\n",
"_____no_output_____"
]
],
[
[
"params = list(net.parameters())\nprint(len(params))\nprint(params[0].size()) # conv1's .weight",
"_____no_output_____"
]
],
[
[
"Let's try a random 32x32 input.\nNote: expected input size of this net (LeNet) is 32x32. To use this net on\nthe MNIST dataset, please resize the images from the dataset to 32x32.\n\n",
"_____no_output_____"
]
],
[
[
"input = torch.randn(1, 1, 32, 32)\nout = net(input)\nprint(out)",
"_____no_output_____"
]
],
[
[
"Zero the gradient buffers of all parameters and backprops with random\ngradients:\n\n",
"_____no_output_____"
]
],
[
[
"net.zero_grad()\nout.backward(torch.randn(1, 10))",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\"><h4>Note</h4><p>``torch.nn`` only supports mini-batches. The entire ``torch.nn``\n package only supports inputs that are a mini-batch of samples, and not\n a single sample.\n\n For example, ``nn.Conv2d`` will take in a 4D Tensor of\n ``nSamples x nChannels x Height x Width``.\n\n If you have a single sample, just use ``input.unsqueeze(0)`` to add\n a fake batch dimension.</p></div>\n\nBefore proceeding further, let's recap all the classes you’ve seen so far.\n\n**Recap:**\n - ``torch.Tensor`` - A *multi-dimensional array* with support for autograd\n operations like ``backward()``. Also *holds the gradient* w.r.t. the\n tensor.\n - ``nn.Module`` - Neural network module. *Convenient way of\n encapsulating parameters*, with helpers for moving them to GPU,\n exporting, loading, etc.\n - ``nn.Parameter`` - A kind of Tensor, that is *automatically\n registered as a parameter when assigned as an attribute to a*\n ``Module``.\n - ``autograd.Function`` - Implements *forward and backward definitions\n of an autograd operation*. Every ``Tensor`` operation creates at\n least a single ``Function`` node that connects to functions that\n created a ``Tensor`` and *encodes its history*.\n\n**At this point, we covered:**\n - Defining a neural network\n - Processing inputs and calling backward\n\n**Still Left:**\n - Computing the loss\n - Updating the weights of the network\n\nLoss Function\n-------------\nA loss function takes the (output, target) pair of inputs, and computes a\nvalue that estimates how far away the output is from the target.\n\nThere are several different\n`loss functions <https://pytorch.org/docs/nn.html#loss-functions>`_ under the\nnn package .\nA simple loss is: ``nn.MSELoss`` which computes the mean-squared error\nbetween the input and the target.\n\nFor example:\n\n",
"_____no_output_____"
]
],
[
[
"output = net(input)\ntarget = torch.randn(10) # a dummy target, for example\ntarget = target.view(1, -1) # make it the same shape as output\ncriterion = nn.MSELoss()\n\nloss = criterion(output, target)\nprint(loss)",
"_____no_output_____"
]
],
[
[
"Now, if you follow ``loss`` in the backward direction, using its\n``.grad_fn`` attribute, you will see a graph of computations that looks\nlike this:\n\n::\n\n input -> conv2d -> relu -> maxpool2d -> conv2d -> relu -> maxpool2d\n -> flatten -> linear -> relu -> linear -> relu -> linear\n -> MSELoss\n -> loss\n\nSo, when we call ``loss.backward()``, the whole graph is differentiated\nw.r.t. the neural net parameters, and all Tensors in the graph that have\n``requires_grad=True`` will have their ``.grad`` Tensor accumulated with the\ngradient.\n\nFor illustration, let us follow a few steps backward:\n\n",
"_____no_output_____"
]
],
[
[
"print(loss.grad_fn) # MSELoss\nprint(loss.grad_fn.next_functions[0][0]) # Linear\nprint(loss.grad_fn.next_functions[0][0].next_functions[0][0]) # ReLU",
"_____no_output_____"
]
],
[
[
"Backprop\n--------\nTo backpropagate the error all we have to do is to ``loss.backward()``.\nYou need to clear the existing gradients though, else gradients will be\naccumulated to existing gradients.\n\n\nNow we shall call ``loss.backward()``, and have a look at conv1's bias\ngradients before and after the backward.\n\n",
"_____no_output_____"
]
],
[
[
"net.zero_grad() # zeroes the gradient buffers of all parameters\n\nprint('conv1.bias.grad before backward')\nprint(net.conv1.bias.grad)\n\nloss.backward()\n\nprint('conv1.bias.grad after backward')\nprint(net.conv1.bias.grad)",
"_____no_output_____"
]
],
[
[
"Now, we have seen how to use loss functions.\n\n**Read Later:**\n\n The neural network package contains various modules and loss functions\n that form the building blocks of deep neural networks. A full list with\n documentation is `here <https://pytorch.org/docs/nn>`_.\n\n**The only thing left to learn is:**\n\n - Updating the weights of the network\n\nUpdate the weights\n------------------\nThe simplest update rule used in practice is the Stochastic Gradient\nDescent (SGD):\n\n ``weight = weight - learning_rate * gradient``\n\nWe can implement this using simple Python code:\n\n.. code:: python\n\n learning_rate = 0.01\n for f in net.parameters():\n f.data.sub_(f.grad.data * learning_rate)\n\nHowever, as you use neural networks, you want to use various different\nupdate rules such as SGD, Nesterov-SGD, Adam, RMSProp, etc.\nTo enable this, we built a small package: ``torch.optim`` that\nimplements all these methods. Using it is very simple:\n\n",
"_____no_output_____"
]
],
[
[
"import torch.optim as optim\n\n# create your optimizer\noptimizer = optim.SGD(net.parameters(), lr=0.01)\n\n# in your training loop:\noptimizer.zero_grad() # zero the gradient buffers\noutput = net(input)\nloss = criterion(output, target)\nloss.backward()\noptimizer.step() # Does the update",
"_____no_output_____"
]
],
[
[
".. Note::\n\n Observe how gradient buffers had to be manually set to zero using\n ``optimizer.zero_grad()``. This is because gradients are accumulated\n as explained in the `Backprop`_ section.\n\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d003223be9b1338421d16f8df58c64c170ca07b7 | 37,255 | ipynb | Jupyter Notebook | intro-to-pytorch/Part 4 - Fashion-MNIST (Exercises).ipynb | xxiMiaxx/deep-learning-v2-pytorch | 0efbd54935c8d0bf214e01627da973f260f5bc90 | [
"MIT"
] | null | null | null | intro-to-pytorch/Part 4 - Fashion-MNIST (Exercises).ipynb | xxiMiaxx/deep-learning-v2-pytorch | 0efbd54935c8d0bf214e01627da973f260f5bc90 | [
"MIT"
] | null | null | null | intro-to-pytorch/Part 4 - Fashion-MNIST (Exercises).ipynb | xxiMiaxx/deep-learning-v2-pytorch | 0efbd54935c8d0bf214e01627da973f260f5bc90 | [
"MIT"
] | null | null | null | 97.782152 | 25,084 | 0.836317 | [
[
[
"# Classifying Fashion-MNIST\n\nNow it's your turn to build and train a neural network. You'll be using the [Fashion-MNIST dataset](https://github.com/zalandoresearch/fashion-mnist), a drop-in replacement for the MNIST dataset. MNIST is actually quite trivial with neural networks where you can easily achieve better than 97% accuracy. Fashion-MNIST is a set of 28x28 greyscale images of clothes. It's more complex than MNIST, so it's a better representation of the actual performance of your network, and a better representation of datasets you'll use in the real world.\n\n<img src='assets/fashion-mnist-sprite.png' width=500px>\n\nIn this notebook, you'll build your own neural network. For the most part, you could just copy and paste the code from Part 3, but you wouldn't be learning. It's important for you to write the code yourself and get it to work. Feel free to consult the previous notebooks though as you work through this.\n\nFirst off, let's load the dataset through torchvision.",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torchvision import datasets, transforms\nimport helper\n\n# Define a transform to normalize the data\ntransform = transforms.Compose([transforms.ToTensor(),\n transforms.Normalize((0.5,), (0.5,))])\n# Download and load the training data\ntrainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)\n\n# Download and load the test data\ntestset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)\ntestloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)",
"Downloading http://fashion-mnist.s3-website.eu-central-1.amazonaws.com/train-images-idx3-ubyte.gz to /Users/Mia/.pytorch/F_MNIST_data/FashionMNIST/raw/train-images-idx3-ubyte.gz\n"
]
],
[
[
"Here we can see one of the images.",
"_____no_output_____"
]
],
[
[
"image, label = next(iter(trainloader))\nhelper.imshow(image[0,:]);",
"_____no_output_____"
]
],
[
[
"## Building the network\n\nHere you should define your network. As with MNIST, each image is 28x28 which is a total of 784 pixels, and there are 10 classes. You should include at least one hidden layer. We suggest you use ReLU activations for the layers and to return the logits or log-softmax from the forward pass. It's up to you how many layers you add and the size of those layers.",
"_____no_output_____"
]
],
[
[
"from torch import nn, optim\nimport torch.nn.functional as F",
"_____no_output_____"
],
[
"# TODO: Define your network architecture here\nclass Network(nn.Module):\n def __init__(self):\n super().__init__()\n self.fc1 = nn.Linear(784, 256)\n self.fc2 = nn.Linear(256, 128)\n self.fc3 = nn.Linear(128, 64)\n self.fc4 = nn.Linear(64, 10)\n \n def forward(self, x):\n #flatten inputs\n x = x.view(x.shape[0], -1)\n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n x = F.relu(self.fc3(x))\n x = F.log_softmax(self.fc4(x), dim = 1)\n \n return x",
"_____no_output_____"
]
],
[
[
"# Train the network\n\nNow you should create your network and train it. First you'll want to define [the criterion](http://pytorch.org/docs/master/nn.html#loss-functions) ( something like `nn.CrossEntropyLoss`) and [the optimizer](http://pytorch.org/docs/master/optim.html) (typically `optim.SGD` or `optim.Adam`).\n\nThen write the training code. Remember the training pass is a fairly straightforward process:\n\n* Make a forward pass through the network to get the logits \n* Use the logits to calculate the loss\n* Perform a backward pass through the network with `loss.backward()` to calculate the gradients\n* Take a step with the optimizer to update the weights\n\nBy adjusting the hyperparameters (hidden units, learning rate, etc), you should be able to get the training loss below 0.4.",
"_____no_output_____"
]
],
[
[
"# TODO: Create the network, define the criterion and optimizer\nmodel = Network()\ncriterion = nn.NLLLoss()\noptimizer = optim.Adam(model.parameters(), lr=0.003)\n\nmodel",
"_____no_output_____"
],
[
"# TODO: Train the network here\n# TODO: Train the network here\n\nepochs = 5\n\nfor e in range(epochs):\n running_loss = 0\n for images, labels in trainloader:\n log_ps = model(images)\n loss = criterion(log_ps, labels)\n \n ## zero grads reset them\n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n running_loss += loss.item()\n\n\n \n \n else:\n print(\"Epoch: \", e)\n print(f\"Training loss: {running_loss/len(trainloader)}\")",
"Epoch: 0\nTraining loss: 0.30448021257578184\nEpoch: 1\nTraining loss: 0.28709665951189967\nEpoch: 2\nTraining loss: 0.2818120150312559\nEpoch: 3\nTraining loss: 0.27553088494391836\nEpoch: 4\nTraining loss: 0.265154266420172\n"
],
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nimport helper\n\n# Test out your network!\n\ndataiter = iter(testloader)\nimages, labels = dataiter.next()\nimg = images[0]\n# Convert 2D image to 1D vector\nimg = img.resize_(1, 784)\n\n# TODO: Calculate the class probabilities (softmax) for img\nps = torch.exp(model(img))\n\n# Plot the image and probabilities\nhelper.view_classify(img.resize_(1, 28, 28), ps, version='Fashion')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d00326101cd523b972639492978d90ab95e4af63 | 4,295 | ipynb | Jupyter Notebook | 1) Download dataset, create .pt files.ipynb | brunoklaus/PS-001-ML5G-GNNetworkingChallenge2021-PARANA | c37abbab19f05388b1ac61a3cc287ff36bdd0e19 | [
"MIT"
] | 7 | 2021-11-04T01:29:10.000Z | 2022-01-20T11:19:55.000Z | 1) Download dataset, create .pt files.ipynb | ITU-AI-ML-in-5G-Challenge/ITU-ML5G-PS-001-PARANA | 137fc23ff8580132f9899657da218d67fec1756c | [
"MIT"
] | null | null | null | 1) Download dataset, create .pt files.ipynb | ITU-AI-ML-in-5G-Challenge/ITU-ML5G-PS-001-PARANA | 137fc23ff8580132f9899657da218d67fec1756c | [
"MIT"
] | null | null | null | 32.537879 | 166 | 0.541327 | [
[
[
"-----------------\n### Please run the IPython Widget below. Using the checkboxes, you can:\n* Download the training, validation and test datasets\n* Extract all tarfiles\n* Create the necessary PyTorch files for the training/validation/test datasets. We create 1 file for each datanet sample, resulting in exactly\n * ./dataset/converted_train/: <b>120,000</b> .pt files <b>(~29.9 GB)</b> \n * ./dataset/converted_val/: <b>3,120</b> .pt files <b>(~14.0 GB)</b> \n * ./dataset/converted_test/: <b>1,560</b> .pt files <b>(~6.7 GB)</b> \n* You can select how many processes to use. Default is 4. More processes = faster runtime due to parallelism, but also multiplies the amount of RAM utilized.\n* Downloaded .gz files are not deleted, free these up manually if you need some space\n-------------------------------------------------------------",
"_____no_output_____"
]
],
[
[
"from convertDataset import process_in_parallel, download_dataset, extract_tarfiles\n\nimport ipywidgets as widgets\ncbs = [widgets.Checkbox() for i in range(5)]\ncbs[0].description=\"Download dataset\"\ncbs[1].description=\"Extract Tarfiles\"\ncbs[2].description=\"Generate Pytorch Files - Training\"\ncbs[3].description=\"Generate Pytorch Files - Validation\"\ncbs[4].description=\"Generate Pytorch Files - Test\"\n\nsl = widgets.IntSlider(\n value=4,\n min=0,\n max=16,\n step=1,\n style= {'description_width': 'initial'},\n layout=widgets.Layout(width='100%',height='80px'),\n description='#processes to use (higher = more parallelism, uses up more RAM)',\n disabled=False,\n continuous_update=False,\n orientation='horizontal',\n readout=True,\n readout_format='d'\n)\n\npb = widgets.Button(\n description='Run',\n disabled=False,\n button_style='', # 'success', 'info', 'warning', 'danger' or ''\n tooltip='Run',\n)\n\ndef on_button_clicked(b):\n if cbs[0].value:\n print(\"Downloading dataset...\")\n download_dataset()\n if cbs[1].value:\n print(\"Extracting Tarfiles...\")\n extract_tarfiles()\n if cbs[2].value:\n print(\"Creating pytorch files (training)...\")\n process_in_parallel('train',sl.value)\n if cbs[3].value:\n print(\"Creating pytorch files (validation)...\")\n process_in_parallel('validation',sl.value)\n if cbs[4].value:\n print(\"Creating pytorch files (test)...\")\n process_in_parallel('test',sl.value)\n\n\npb.on_click(on_button_clicked)\n\nui = widgets.VBox([widgets.HBox([x]) for x in cbs+[sl]] +[pb])\n\ndisplay(ui)\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
d0032d65102a1a810d0135cd280d208c57458812 | 14,124 | ipynb | Jupyter Notebook | notebooks/figure_supervised_comp.ipynb | priyaravichander/ganspace | fefc001fabf6986a98da4df3166fc31693a4c26b | [
"Apache-2.0"
] | 1,644 | 2020-04-07T01:00:10.000Z | 2022-03-30T10:27:13.000Z | notebooks/figure_supervised_comp.ipynb | priyaravichander/ganspace | fefc001fabf6986a98da4df3166fc31693a4c26b | [
"Apache-2.0"
] | 54 | 2020-04-07T23:32:19.000Z | 2022-03-27T15:06:26.000Z | notebooks/figure_supervised_comp.ipynb | priyaravichander/ganspace | fefc001fabf6986a98da4df3166fc31693a4c26b | [
"Apache-2.0"
] | 224 | 2020-04-06T22:59:44.000Z | 2022-03-29T14:35:45.000Z | 40.239316 | 171 | 0.585245 | [
[
[
"# Copyright 2020 Erik Härkönen. All rights reserved.\n# This file is licensed to you under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License. You may obtain a copy\n# of the License at http://www.apache.org/licenses/LICENSE-2.0\n\n# Unless required by applicable law or agreed to in writing, software distributed under\n# the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR REPRESENTATIONS\n# OF ANY KIND, either express or implied. See the License for the specific language\n# governing permissions and limitations under the License.\n\n# Comparison to GAN steerability and InterfaceGAN\n%matplotlib inline\nfrom notebook_init import *\nimport pickle\n\nout_root = Path('out/figures/steerability_comp')\nmakedirs(out_root, exist_ok=True)\nrand = lambda : np.random.randint(np.iinfo(np.int32).max)",
"_____no_output_____"
],
[
"def show_strip(frames):\n plt.figure(figsize=(20,20))\n plt.axis('off')\n plt.imshow(np.hstack(pad_frames(frames, 64)))\n plt.show()",
"_____no_output_____"
],
[
"normalize = lambda t : t / np.sqrt(np.sum(t.reshape(-1)**2))\n\ndef compute(\n model,\n lat_mean,\n prefix,\n imgclass,\n seeds,\n d_ours,\n l_start,\n l_end,\n scale_ours,\n d_sup, # single or one per layer\n scale_sup,\n center=True\n):\n model.set_output_class(imgclass)\n makedirs(out_root / imgclass, exist_ok=True)\n \n for seed in seeds:\n print(seed)\n deltas = [d_ours, d_sup]\n scales = [scale_ours, scale_sup]\n ranges = [(l_start, l_end), (0, model.get_max_latents())]\n names = ['ours', 'supervised']\n\n for delta, name, scale, l_range in zip(deltas, names, scales, ranges):\n lat_base = model.sample_latent(1, seed=seed).cpu().numpy()\n\n # Shift latent to lie on mean along given direction\n if center:\n y = normalize(d_sup) # assume ground truth\n dotp = np.sum((lat_base - lat_mean) * y, axis=-1, keepdims=True)\n lat_base = lat_base - dotp * y\n \n # Convert single delta to per-layer delta (to support Steerability StyleGAN)\n if delta.shape[0] > 1:\n #print('Unstacking delta')\n *d_per_layer, = delta # might have per-layer scales, don't normalize\n else:\n d_per_layer = [normalize(delta)]*model.get_max_latents()\n \n frames = []\n n_frames = 5\n for a in np.linspace(-1.0, 1.0, n_frames):\n w = [lat_base]*model.get_max_latents()\n for l in range(l_range[0], l_range[1]):\n w[l] = w[l] + a*d_per_layer[l]*scale\n frames.append(model.sample_np(w))\n\n for i, frame in enumerate(frames):\n Image.fromarray(np.uint8(frame*255)).save(\n out_root / imgclass / f'{prefix}_{name}_{seed}_{i}.png')\n \n strip = np.hstack(pad_frames(frames, 64))\n plt.figure(figsize=(12,12))\n plt.imshow(strip)\n plt.axis('off')\n plt.tight_layout()\n plt.title(f'{prefix} - {name}, scale={scale}')\n plt.show()",
"_____no_output_____"
],
[
"# BigGAN-512\n\ninst = get_instrumented_model('BigGAN-512', 'husky', 'generator.gen_z', device, inst=inst)\nmodel = inst.model\n\nK = model.get_max_latents()\npc_config = Config(components=128, n=1_000_000,\n layer='generator.gen_z', model='BigGAN-512', output_class='husky')\ndump_name = get_or_compute(pc_config, inst)\n\nwith np.load(dump_name) as data:\n lat_comp = data['lat_comp']\n lat_mean = data['lat_mean']\n\nwith open('data/steerability/biggan_deep_512/gan_steer-linear_zoom_512.pkl', 'rb') as f:\n delta_steerability_zoom = pickle.load(f)['w_zoom'].reshape(1, 128)\nwith open('data/steerability/biggan_deep_512/gan_steer-linear_shiftx_512.pkl', 'rb') as f:\n delta_steerability_transl = pickle.load(f)['w_shiftx'].reshape(1, 128)\n\n# Indices determined by visual inspection\ndelta_ours_transl = lat_comp[0]\ndelta_ours_zoom = lat_comp[6]\n\nmodel.truncation = 0.6\ncompute(model, lat_mean, 'zoom', 'robin', [560157313], delta_ours_zoom, 0, K, -3.0, delta_steerability_zoom, 5.5)\ncompute(model, lat_mean, 'zoom', 'ship', [107715983], delta_ours_zoom, 0, K, -3.0, delta_steerability_zoom, 5.0)\n\ncompute(model, lat_mean, 'translate', 'golden_retriever', [552411435], delta_ours_transl, 0, K, -2.0, delta_steerability_transl, 4.5)\ncompute(model, lat_mean, 'translate', 'lemon', [331582800], delta_ours_transl, 0, K, -3.0, delta_steerability_transl, 6.0)",
"_____no_output_____"
],
[
"# StyleGAN1-ffhq (InterfaceGAN)\n\ninst = get_instrumented_model('StyleGAN', 'ffhq', 'g_mapping', device, use_w=True, inst=inst)\nmodel = inst.model\n\nK = model.get_max_latents()\npc_config = Config(components=128, n=1_000_000, use_w=True,\n layer='g_mapping', model='StyleGAN', output_class='ffhq')\ndump_name = get_or_compute(pc_config, inst)\n\nwith np.load(dump_name) as data:\n lat_comp = data['lat_comp']\n lat_mean = data['lat_mean']\n\n# SG-ffhq-w, non-conditional\nd_ffhq_pose = np.load('data/interfacegan/stylegan_ffhq_pose_w_boundary.npy').astype(np.float32)\nd_ffhq_smile = np.load('data/interfacegan/stylegan_ffhq_smile_w_boundary.npy').astype(np.float32)\nd_ffhq_gender = np.load('data/interfacegan/stylegan_ffhq_gender_w_boundary.npy').astype(np.float32)\nd_ffhq_glasses = np.load('data/interfacegan/stylegan_ffhq_eyeglasses_w_boundary.npy').astype(np.float32)\n\n# Indices determined by visual inspection\nd_ours_pose = lat_comp[9]\nd_ours_smile = lat_comp[44]\nd_ours_gender = lat_comp[0]\nd_ours_glasses = lat_comp[12]\n\nmodel.truncation = 1.0 # NOT IMPLEMENTED\ncompute(model, lat_mean, 'pose', 'ffhq', [440608316, 1811098088, 129888612], d_ours_pose, 0, 7, -1.0, d_ffhq_pose, 1.0)\ncompute(model, lat_mean, 'smile', 'ffhq', [1759734403, 1647189561, 70163682], d_ours_smile, 3, 4, -8.5, d_ffhq_smile, 1.0)\ncompute(model, lat_mean, 'gender', 'ffhq', [1302836080, 1746672325], d_ours_gender, 2, 6, -4.5, d_ffhq_gender, 1.5)\ncompute(model, lat_mean, 'glasses', 'ffhq', [1565213752, 1005764659, 1110182583], d_ours_glasses, 0, 2, 4.0, d_ffhq_glasses, 1.0)",
"_____no_output_____"
],
[
"# StyleGAN1-ffhq (Steerability)\n\ninst = get_instrumented_model('StyleGAN', 'ffhq', 'g_mapping', device, use_w=True, inst=inst)\nmodel = inst.model\n\nK = model.get_max_latents()\npc_config = Config(components=128, n=1_000_000, use_w=True,\n layer='g_mapping', model='StyleGAN', output_class='ffhq')\ndump_name = get_or_compute(pc_config, inst)\n\nwith np.load(dump_name) as data:\n lat_comp = data['lat_comp']\n lat_mean = data['lat_mean']\n\n# SG-ffhq-w, non-conditional\n# Shapes: [18, 512]\nd_ffhq_R = np.load('data/steerability/stylegan_ffhq/ffhq_rgb_0.npy').astype(np.float32)\nd_ffhq_G = np.load('data/steerability/stylegan_ffhq/ffhq_rgb_1.npy').astype(np.float32)\nd_ffhq_B = np.load('data/steerability/stylegan_ffhq/ffhq_rgb_2.npy').astype(np.float32)\n\n# Indices determined by visual inspection\nd_ours_R = lat_comp[0]\nd_ours_G = -lat_comp[1]\nd_ours_B = -lat_comp[2]\n\nmodel.truncation = 1.0 # NOT IMPLEMENTED\ncompute(model, lat_mean, 'red', 'ffhq', [5], d_ours_R, 17, 18, 8.0, d_ffhq_R, 1.0, center=False)\ncompute(model, lat_mean, 'green', 'ffhq', [5], d_ours_G, 17, 18, 15.0, d_ffhq_G, 1.0, center=False)\ncompute(model, lat_mean, 'blue', 'ffhq', [5], d_ours_B, 17, 18, 10.0, d_ffhq_B, 1.0, center=False)",
"_____no_output_____"
],
[
"# StyleGAN1-celebahq (InterfaceGAN)\n\ninst = get_instrumented_model('StyleGAN', 'celebahq', 'g_mapping', device, use_w=True, inst=inst)\nmodel = inst.model\n\nK = model.get_max_latents()\npc_config = Config(components=128, n=1_000_000, use_w=True,\n layer='g_mapping', model='StyleGAN', output_class='celebahq')\ndump_name = get_or_compute(pc_config, inst)\n\nwith np.load(dump_name) as data:\n lat_comp = data['lat_comp']\n lat_mean = data['lat_mean']\n\n# SG-ffhq-w, non-conditional\nd_celebahq_pose = np.load('data/interfacegan/stylegan_celebahq_pose_w_boundary.npy').astype(np.float32)\nd_celebahq_smile = np.load('data/interfacegan/stylegan_celebahq_smile_w_boundary.npy').astype(np.float32)\nd_celebahq_gender = np.load('data/interfacegan/stylegan_celebahq_gender_w_boundary.npy').astype(np.float32)\nd_celebahq_glasses = np.load('data/interfacegan/stylegan_celebahq_eyeglasses_w_boundary.npy').astype(np.float32)\n\n# Indices determined by visual inspection\nd_ours_pose = lat_comp[7]\nd_ours_smile = lat_comp[14]\nd_ours_gender = lat_comp[1]\nd_ours_glasses = lat_comp[5]\n\nmodel.truncation = 1.0 # NOT IMPLEMENTED\ncompute(model, lat_mean, 'pose', 'celebahq', [1939067252, 1460055449, 329555154], d_ours_pose, 0, 7, -1.0, d_celebahq_pose, 1.0)\ncompute(model, lat_mean, 'smile', 'celebahq', [329187806, 424805522, 1777796971], d_ours_smile, 3, 4, -7.0, d_celebahq_smile, 1.3)\ncompute(model, lat_mean, 'gender', 'celebahq', [1144615644, 967075839, 264878205], d_ours_gender, 0, 2, -3.2, d_celebahq_gender, 1.2)\ncompute(model, lat_mean, 'glasses', 'celebahq', [991993380, 594344173, 2119328990, 1919124025], d_ours_glasses, 0, 1, -10.0, d_celebahq_glasses, 2.0) # hard for both",
"_____no_output_____"
],
[
"# StyleGAN1-cars (Steerability)\n\ninst = get_instrumented_model('StyleGAN', 'cars', 'g_mapping', device, use_w=True, inst=inst)\nmodel = inst.model\n\nK = model.get_max_latents()\npc_config = Config(components=128, n=1_000_000, use_w=True,\n layer='g_mapping', model='StyleGAN', output_class='cars')\ndump_name = get_or_compute(pc_config, inst)\n\nwith np.load(dump_name) as data:\n lat_comp = data['lat_comp']\n lat_mean = data['lat_mean']\n\n# Shapes: [16, 512]\nd_cars_rot = np.load('data/steerability/stylegan_cars/rotate2d.npy').astype(np.float32)\nd_cars_shift = np.load('data/steerability/stylegan_cars/shifty.npy').astype(np.float32)\n\n# Add two final layers\nd_cars_rot = np.append(d_cars_rot, np.zeros((2,512), dtype=np.float32), axis=0)\nd_cars_shift = np.append(d_cars_shift, np.zeros((2,512), dtype=np.float32), axis=0)\n\nprint(d_cars_rot.shape)\n\n# Indices determined by visual inspection\nd_ours_rot = lat_comp[0]\nd_ours_shift = lat_comp[7]\n\nmodel.truncation = 1.0 # NOT IMPLEMENTED\ncompute(model, lat_mean, 'rotate2d', 'cars', [46, 28], d_ours_rot, 0, 1, 1.0, d_cars_rot, 1.0, center=False)\ncompute(model, lat_mean, 'shifty', 'cars', [0, 13], d_ours_shift, 1, 2, 4.0, d_cars_shift, 1.0, center=False)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0034efb56063d527ae231c92775c2b06c118e2a | 311,039 | ipynb | Jupyter Notebook | Untitled.ipynb | gaben3722/Time-Series-Project | 44186635e7d2f62648cc9aad4154f4003bd397c4 | [
"MIT"
] | null | null | null | Untitled.ipynb | gaben3722/Time-Series-Project | 44186635e7d2f62648cc9aad4154f4003bd397c4 | [
"MIT"
] | null | null | null | Untitled.ipynb | gaben3722/Time-Series-Project | 44186635e7d2f62648cc9aad4154f4003bd397c4 | [
"MIT"
] | null | null | null | 505.754472 | 116,144 | 0.942705 | [
[
[
"# Importing Dependencies",
"_____no_output_____"
]
],
[
[
"import numpy as np \nimport pandas as pd \nimport matplotlib.pyplot as plt \nimport seaborn as sns\nimport pandas_datareader\nimport pandas_datareader.data as web\nimport datetime\nfrom sklearn.preprocessing import MinMaxScaler\nfrom keras.models import Sequential\nfrom keras.layers import Dense,LSTM,Dropout\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# Importing Data",
"_____no_output_____"
]
],
[
[
"start = datetime.datetime(2016,1,1)\nend = datetime.datetime(2021,1,1)",
"_____no_output_____"
],
[
"QQQ = web.DataReader(\"QQQ\", \"yahoo\", start, end)\nQQQ.head()",
"_____no_output_____"
],
[
"QQQ['Close'].plot(label = 'QQQ', figsize = (16,10), title = 'Closing Price')\nplt.legend();",
"_____no_output_____"
],
[
"QQQ['Volume'].plot(label = 'QQQ', figsize = (16,10), title = 'Volume Traded')\nplt.legend();",
"_____no_output_____"
],
[
"QQQ['MA50'] = QQQ['Close'].rolling(50).mean()\nQQQ['MA200'] = QQQ['Close'].rolling(200).mean()\nQQQ[['Close','MA50','MA200']].plot(figsize = (16,10), title = 'Moving Averages')",
"_____no_output_____"
]
],
[
[
"# Selecting The Close Column",
"_____no_output_____"
]
],
[
[
"QQQ[\"Close\"]=pd.to_numeric(QQQ.Close,errors='coerce') #turning the Close column to numeric\nQQQ = QQQ.dropna() \ntrainData = QQQ.iloc[:,3:4].values #selecting closing prices for training",
"_____no_output_____"
]
],
[
[
"# Scaling Values in the Range of 0-1 for Best Results",
"_____no_output_____"
]
],
[
[
"sc = MinMaxScaler(feature_range=(0,1))\ntrainData = sc.fit_transform(trainData)\ntrainData.shape",
"_____no_output_____"
]
],
[
[
"# Prepping Data for LSTM",
"_____no_output_____"
]
],
[
[
"X_train = []\ny_train = []\n\nfor i in range (60,1060): \n X_train.append(trainData[i-60:i,0]) \n y_train.append(trainData[i,0])\n\nX_train,y_train = np.array(X_train),np.array(y_train)",
"_____no_output_____"
],
[
"X_train = np.reshape(X_train,(X_train.shape[0],X_train.shape[1],1)) #adding the batch_size axis\nX_train.shape",
"_____no_output_____"
]
],
[
[
"# Building The Model",
"_____no_output_____"
]
],
[
[
"model = Sequential()\n\nmodel.add(LSTM(units=100, return_sequences = True, input_shape =(X_train.shape[1],1)))\nmodel.add(Dropout(0.2))\n\nmodel.add(LSTM(units=100, return_sequences = True))\nmodel.add(Dropout(0.2))\n\nmodel.add(LSTM(units=100, return_sequences = True))\nmodel.add(Dropout(0.2))\n\nmodel.add(LSTM(units=100, return_sequences = False))\nmodel.add(Dropout(0.2))\n\nmodel.add(Dense(units =1))\nmodel.compile(optimizer='adam',loss=\"mean_squared_error\")",
"_____no_output_____"
],
[
"hist = model.fit(X_train, y_train, epochs = 20, batch_size = 32, verbose=2)",
"Epoch 1/20\n32/32 - 26s - loss: 0.0187\nEpoch 2/20\n32/32 - 3s - loss: 0.0036\nEpoch 3/20\n32/32 - 3s - loss: 0.0026\nEpoch 4/20\n32/32 - 3s - loss: 0.0033\nEpoch 5/20\n32/32 - 3s - loss: 0.0033\nEpoch 6/20\n32/32 - 3s - loss: 0.0028\nEpoch 7/20\n32/32 - 3s - loss: 0.0024\nEpoch 8/20\n32/32 - 3s - loss: 0.0024\nEpoch 9/20\n32/32 - 3s - loss: 0.0030\nEpoch 10/20\n32/32 - 3s - loss: 0.0026\nEpoch 11/20\n32/32 - 3s - loss: 0.0020\nEpoch 12/20\n32/32 - 3s - loss: 0.0018\nEpoch 13/20\n32/32 - 3s - loss: 0.0024\nEpoch 14/20\n32/32 - 3s - loss: 0.0020\nEpoch 15/20\n32/32 - 3s - loss: 0.0029\nEpoch 16/20\n32/32 - 4s - loss: 0.0022\nEpoch 17/20\n32/32 - 3s - loss: 0.0016\nEpoch 18/20\n32/32 - 3s - loss: 0.0029\nEpoch 19/20\n32/32 - 3s - loss: 0.0021\nEpoch 20/20\n32/32 - 3s - loss: 0.0015\n"
]
],
[
[
"# Plotting The Training Loss",
"_____no_output_____"
]
],
[
[
"plt.plot(hist.history['loss'])\nplt.title('model loss')\nplt.ylabel('loss')\nplt.xlabel('epoch')\nplt.legend(['train'], loc='upper left')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Testing Model on New Data",
"_____no_output_____"
]
],
[
[
"start = datetime.datetime(2021,1,1) \nend = datetime.datetime.today()",
"_____no_output_____"
],
[
"testData = web.DataReader(\"QQQ\", \"yahoo\", start, end) #importing new data for testing\ntestData[\"Close\"]=pd.to_numeric(testData.Close,errors='coerce') #turning the Close column to numeric\ntestData = testData.dropna() #droping the NA values\ntestData = testData.iloc[:,3:4] #selecting the closing prices for testing\ny_test = testData.iloc[60:,0:].values #selecting the labels \n#input array for the model\ninputClosing = testData.iloc[:,0:].values \ninputClosing_scaled = sc.transform(inputClosing)\ninputClosing_scaled.shape\nX_test = []\nlength = len(testData)\ntimestep = 60\nfor i in range(timestep,length): \n X_test.append(inputClosing_scaled[i-timestep:i,0])\nX_test = np.array(X_test)\nX_test = np.reshape(X_test,(X_test.shape[0],X_test.shape[1],1))\nX_test.shape",
"_____no_output_____"
],
[
"y_pred = model.predict(X_test) #predicting values",
"_____no_output_____"
],
[
"predicted_price = sc.inverse_transform(y_pred) #inversing the scaling transformation for plotting ",
"_____no_output_____"
]
],
[
[
"# Plotting Results",
"_____no_output_____"
]
],
[
[
"plt.plot(y_test, color = 'blue', label = 'Actual Stock Price')\nplt.plot(predicted_price, color = 'red', label = 'Predicted Stock Price')\nplt.title('QQQ stock price prediction')\nplt.xlabel('Time')\nplt.ylabel('Stock Price')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d003503005a19dc652ce59c887923c0aa6c5c0eb | 9,739 | ipynb | Jupyter Notebook | Expressions and Operations.ipynb | CedricPengson/CPEN-21-A-ECE-2-1 | cf5d5a43b2d66da6e520614620dea75e47e8d85f | [
"Apache-2.0"
] | null | null | null | Expressions and Operations.ipynb | CedricPengson/CPEN-21-A-ECE-2-1 | cf5d5a43b2d66da6e520614620dea75e47e8d85f | [
"Apache-2.0"
] | null | null | null | Expressions and Operations.ipynb | CedricPengson/CPEN-21-A-ECE-2-1 | cf5d5a43b2d66da6e520614620dea75e47e8d85f | [
"Apache-2.0"
] | null | null | null | 21.787472 | 258 | 0.384947 | [
[
[
"<a href=\"https://colab.research.google.com/github/CedricPengson/CPEN-21-A-ECE-2-1/blob/main/Expressions%20and%20Operations.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"##Boolean Operator\n",
"_____no_output_____"
]
],
[
[
"print (10>9)\nprint (10==9)\nprint (10<9)",
"True\nFalse\nFalse\n"
],
[
"x=1\ny=2\nprint(x>y)\nprint(10>11)\nprint(10==10)\nprint(10!=11)",
"False\nFalse\nTrue\nTrue\n"
],
[
"#using bool() function\n\nprint(bool(\"Hello\"))\nprint(bool(15))\nprint(bool(1))\n\nprint(bool(True))\nprint(bool(False))\nprint(bool(0))\nprint(bool([]))\n",
"True\nTrue\nTrue\nTrue\nFalse\nFalse\nFalse\n"
]
],
[
[
"##Functions can return Boolean",
"_____no_output_____"
]
],
[
[
"def myfunctionboolean(): return True\nprint(myfunctionboolean())\n",
"True\n"
],
[
"def myfunction(): return False\n\nif myfunction():\n print(\"yes!\")\nelse:\n print(\"no\")",
"no\n"
]
],
[
[
"##You Try",
"_____no_output_____"
]
],
[
[
"print(10>9)\n\na=6\nb=7\nprint(a==b)\nprint(a!=a)\n",
"True\nFalse\nFalse\n"
]
],
[
[
"##Arithmetic Operators",
"_____no_output_____"
]
],
[
[
"print(10+5)\nprint(10-5)\nprint(10*5)\nprint(10/5)\n\nprint(10%5) #modulo division, remainder\nprint(10//5) #floor division\nprint(10//3) #floor division\nprint(10%3) #3x3=9+1\n\nprint(10**5)\n\n",
"15\n5\n50\n2.0\n0\n2\n3\n1\n100000\n"
]
],
[
[
"##Bitwise Operators",
"_____no_output_____"
]
],
[
[
"a=60 #0011 1100\nb=13 #0000 1101\n\nprint(a&b)\nprint(a|b)\nprint(a^b)\nprint(~a)\nprint(a<<1) #0111 1000\nprint(a<<2) #1111 0000\nprint(b>>1) #1 0000 0110\nprint(b>>2) #0000 0011 carry flag bit=01 ",
"12\n61\n49\n-61\n120\n240\n6\n3\n"
]
],
[
[
"##Phyton Assigment Operators",
"_____no_output_____"
]
],
[
[
"a+=3 #Same As a = a + 3\n #Same As a = 60 + 3, a=63\nprint(a)",
"63\n"
]
],
[
[
"##Logical Operators",
"_____no_output_____"
]
],
[
[
"#and logical operators\n\na = True\nb = False\n\nprint(a and b)\nprint(not(a and b))\nprint(a or b)\nprint(not(a or b))",
"False\nTrue\nTrue\nFalse\n"
],
[
"print(a is b)\nprint(a is not b)",
"False\nTrue\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d00352c042e11023e04cd767f979253bf98e6a8d | 11,488 | ipynb | Jupyter Notebook | Vine_Review_Analysis.ipynb | ethiry99/HW16_Amazon_Vine_Analysis | efbd4d44125888472f833ff8c3304848796caa7a | [
"MIT"
] | null | null | null | Vine_Review_Analysis.ipynb | ethiry99/HW16_Amazon_Vine_Analysis | efbd4d44125888472f833ff8c3304848796caa7a | [
"MIT"
] | null | null | null | Vine_Review_Analysis.ipynb | ethiry99/HW16_Amazon_Vine_Analysis | efbd4d44125888472f833ff8c3304848796caa7a | [
"MIT"
] | null | null | null | 25.93228 | 156 | 0.409384 | [
[
[
"# Dependencies and Setup\nimport pandas as pd",
"_____no_output_____"
],
[
"vine_review_df=pd.read_csv(\"Resources/vine_table.csv\")\n",
"_____no_output_____"
],
[
"vine_review_df.head()\n",
"_____no_output_____"
],
[
"vine_review_df=vine_review_df.loc[(vine_review_df[\"total_votes\"] >= 20) & (vine_review_df[\"helpful_votes\"]/vine_review_df[\"total_votes\"] >= .5)]",
"_____no_output_____"
],
[
"vine_review_df.head()",
"_____no_output_____"
],
[
"vine_rv_paid_df=vine_review_df.loc[vine_review_df[\"vine\"]==\"Y\"]\n\nvine_rv_paid_count=len(vine_rv_paid_df)\n\n#print(f\"5 Star paid percent {vine_five_star_paid_percent:.1%}\\n\"\n\nprint(f\"Paid vine reviews = {vine_rv_paid_count}\")",
"Paid vine reviews = 386\n"
],
[
"vine_rv_unpaid_df=vine_review_df.loc[vine_review_df[\"vine\"]==\"N\"]\n\nvine_rv_unpaid_count=len(vine_rv_unpaid_df)\n\nprint(f\"Paid (vine) reviews = {vine_rv_paid_count}\")\nprint(f\"Unpaid (vine) reviews = {vine_rv_unpaid_count}\")",
"Paid (vine) reviews = 386\nUnpaid (vine) reviews = 48717\n"
],
[
"vine_rv_paid_five_star_df=vine_rv_paid_df.loc[(vine_rv_paid_df[\"star_rating\"]==5)]\nfive_star_paid_count=len(vine_rv_paid_five_star_df)\nprint(f\"Five star paid reviews = {five_star_paid_count}\")",
"Five star paid reviews = 176\n"
],
[
"vine_rv_unpaid_five_star_df=vine_rv_unpaid_df.loc[(vine_rv_unpaid_df[\"star_rating\"]==5)]\nfive_star_unpaid_count=len(vine_rv_unpaid_five_star_df)\nprint(f\"Five star paid reviews = {five_star_paid_count}\")\nprint(f\"Five star unpaid reviews = {five_star_unpaid_count}\")",
"Five star paid reviews = 176\nFive star unpaid reviews = 24026\n"
],
[
"vine_five_star_paid_percent=five_star_paid_count/vine_rv_paid_count\nvine_five_star_paid_percent",
"_____no_output_____"
],
[
"vine_five_star_unpaid_percent=five_star_unpaid_count/vine_rv_unpaid_count\nvine_five_star_unpaid_percent",
"_____no_output_____"
],
[
"print(f\"5 Star paid percent {vine_five_star_paid_percent:.1%}\\n\"\n f\"5 Star unpaid percent {vine_five_star_unpaid_percent:.1%}\")",
"5 Star paid percent 45.6%\n5 Star unpaid percent 49.3%\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d00358ed9a2b3961036e248c0ed61a57653a1f75 | 538 | ipynb | Jupyter Notebook | Python/Scikit-learn.ipynb | JnuLi/DataScience | 3a0c5992a84c1f9633fe7b27c2252f5964cb3f8d | [
"Apache-2.0"
] | null | null | null | Python/Scikit-learn.ipynb | JnuLi/DataScience | 3a0c5992a84c1f9633fe7b27c2252f5964cb3f8d | [
"Apache-2.0"
] | null | null | null | Python/Scikit-learn.ipynb | JnuLi/DataScience | 3a0c5992a84c1f9633fe7b27c2252f5964cb3f8d | [
"Apache-2.0"
] | null | null | null | 16.30303 | 34 | 0.522305 | [
[
[
"# Scikit-learn",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
d0036fb286e2bf06c17074b7ce00dd63c3d694f9 | 81,065 | ipynb | Jupyter Notebook | Coursera/Art and Science of Machine Learning/Improve model accuracy by hyperparameter tuning with AI Platform.ipynb | helpthx/Path_through_Data_Science_2019 | aa22333eae970506f2ce184551c55565b0be89fb | [
"MIT"
] | 2 | 2019-02-06T09:30:44.000Z | 2019-02-09T18:24:46.000Z | Coursera/Art and Science of Machine Learning/Improve model accuracy by hyperparameter tuning with AI Platform.ipynb | helpthx/Path_through_Data_Science_2019 | aa22333eae970506f2ce184551c55565b0be89fb | [
"MIT"
] | 11 | 2019-06-22T00:58:03.000Z | 2019-07-27T14:59:21.000Z | Coursera/Art and Science of Machine Learning/Improve model accuracy by hyperparameter tuning with AI Platform.ipynb | helpthx/Path_through_Data_Science_2019 | aa22333eae970506f2ce184551c55565b0be89fb | [
"MIT"
] | 1 | 2020-12-03T21:10:43.000Z | 2020-12-03T21:10:43.000Z | 76.984805 | 553 | 0.706211 | [
[
[
"# Hyperparameter tuning with Cloud AI Platform",
"_____no_output_____"
],
[
"**Learning Objectives:**\n * Improve the accuracy of a model by hyperparameter tuning",
"_____no_output_____"
]
],
[
[
"import os\nPROJECT = 'qwiklabs-gcp-faf328caac1ef9a0' # REPLACE WITH YOUR PROJECT ID\nBUCKET = 'qwiklabs-gcp-faf328caac1ef9a0' # REPLACE WITH YOUR BUCKET NAME\nREGION = 'us-east1' # REPLACE WITH YOUR BUCKET REGION e.g. us-central1",
"_____no_output_____"
],
[
"# for bash\nos.environ['PROJECT'] = PROJECT\nos.environ['BUCKET'] = BUCKET\nos.environ['REGION'] = REGION\nos.environ['TFVERSION'] = '1.8' # Tensorflow version",
"_____no_output_____"
],
[
"%%bash\ngcloud config set project $PROJECT\ngcloud config set compute/region $REGION",
"Updated property [core/project].\nUpdated property [compute/region].\n"
]
],
[
[
"## Create command-line program\n\nIn order to submit to Cloud AI Platform, we need to create a distributed training program. Let's convert our housing example to fit that paradigm, using the Estimators API.",
"_____no_output_____"
]
],
[
[
"%%bash\nrm -rf house_prediction_module\nmkdir house_prediction_module\nmkdir house_prediction_module/trainer\ntouch house_prediction_module/trainer/__init__.py",
"_____no_output_____"
],
[
"%%writefile house_prediction_module/trainer/task.py\nimport argparse\nimport os\nimport json\nimport shutil\n\nfrom . import model\n \nif __name__ == '__main__' and \"get_ipython\" not in dir():\n parser = argparse.ArgumentParser()\n parser.add_argument(\n '--learning_rate',\n type = float, \n default = 0.01\n )\n parser.add_argument(\n '--batch_size',\n type = int, \n default = 30\n )\n parser.add_argument(\n '--output_dir',\n help = 'GCS location to write checkpoints and export models.',\n required = True\n )\n parser.add_argument(\n '--job-dir',\n help = 'this model ignores this field, but it is required by gcloud',\n default = 'junk'\n )\n args = parser.parse_args()\n arguments = args.__dict__\n \n # Unused args provided by service\n arguments.pop('job_dir', None)\n arguments.pop('job-dir', None)\n \n # Append trial_id to path if we are doing hptuning\n # This code can be removed if you are not using hyperparameter tuning\n arguments['output_dir'] = os.path.join(\n arguments['output_dir'],\n json.loads(\n os.environ.get('TF_CONFIG', '{}')\n ).get('task', {}).get('trial', '')\n )\n \n # Run the training\n shutil.rmtree(arguments['output_dir'], ignore_errors=True) # start fresh each time\n \n # Pass the command line arguments to our model's train_and_evaluate function\n model.train_and_evaluate(arguments)",
"Writing house_prediction_module/trainer/task.py\n"
],
[
"%%writefile house_prediction_module/trainer/model.py\n\nimport numpy as np\nimport pandas as pd\nimport tensorflow as tf\n\ntf.logging.set_verbosity(tf.logging.INFO)\n\n# Read dataset and split into train and eval\ndf = pd.read_csv(\"https://storage.googleapis.com/ml_universities/california_housing_train.csv\", sep = \",\")\ndf['num_rooms'] = df['total_rooms'] / df['households']\nnp.random.seed(seed = 1) #makes split reproducible\nmsk = np.random.rand(len(df)) < 0.8\ntraindf = df[msk]\nevaldf = df[~msk]\n\n# Train and eval input functions\nSCALE = 100000\n\ndef train_input_fn(df, batch_size):\n return tf.estimator.inputs.pandas_input_fn(x = traindf[[\"num_rooms\"]],\n y = traindf[\"median_house_value\"] / SCALE, # note the scaling\n num_epochs = None,\n batch_size = batch_size, # note the batch size\n shuffle = True)\n\ndef eval_input_fn(df, batch_size):\n return tf.estimator.inputs.pandas_input_fn(x = evaldf[[\"num_rooms\"]],\n y = evaldf[\"median_house_value\"] / SCALE, # note the scaling\n num_epochs = 1,\n batch_size = batch_size,\n shuffle = False)\n\n# Define feature columns\nfeatures = [tf.feature_column.numeric_column('num_rooms')]\n\ndef train_and_evaluate(args):\n # Compute appropriate number of steps\n num_steps = (len(traindf) / args['batch_size']) / args['learning_rate'] # if learning_rate=0.01, hundred epochs\n\n # Create custom optimizer\n myopt = tf.train.FtrlOptimizer(learning_rate = args['learning_rate']) # note the learning rate\n\n # Create rest of the estimator as usual\n estimator = tf.estimator.LinearRegressor(model_dir = args['output_dir'], \n feature_columns = features, \n optimizer = myopt)\n #Add rmse evaluation metric\n def rmse(labels, predictions):\n pred_values = tf.cast(predictions['predictions'], tf.float64)\n return {'rmse': tf.metrics.root_mean_squared_error(labels * SCALE, pred_values * SCALE)}\n estimator = tf.contrib.estimator.add_metrics(estimator, rmse)\n\n train_spec = tf.estimator.TrainSpec(input_fn = train_input_fn(df = traindf, batch_size = args['batch_size']),\n max_steps = num_steps)\n eval_spec = tf.estimator.EvalSpec(input_fn = eval_input_fn(df = evaldf, batch_size = len(evaldf)),\n steps = None)\n tf.estimator.train_and_evaluate(estimator, train_spec, eval_spec)",
"Writing house_prediction_module/trainer/model.py\n"
],
[
"%%bash\nrm -rf house_trained\nexport PYTHONPATH=${PYTHONPATH}:${PWD}/house_prediction_module\ngcloud ai-platform local train \\\n --module-name=trainer.task \\\n --job-dir=house_trained \\\n --package-path=$(pwd)/trainer \\\n -- \\\n --batch_size=30 \\\n --learning_rate=0.02 \\\n --output_dir=house_trained",
"WARNING: Logging before flag parsing goes to stderr.\nW0809 20:42:02.240282 139715572925888 deprecation_wrapper.py:119] From /home/jupyter/training-data-analyst/courses/machine_learning/deepdive/05_artandscience/house_prediction_module/trainer/model.py:6: The name tf.logging.set_verbosity is deprecated. Please use tf.compat.v1.logging.set_verbosity instead.\n\nW0809 20:42:02.240634 139715572925888 deprecation_wrapper.py:119] From /home/jupyter/training-data-analyst/courses/machine_learning/deepdive/05_artandscience/house_prediction_module/trainer/model.py:6: The name tf.logging.INFO is deprecated. Please use tf.compat.v1.logging.INFO instead.\n\nW0809 20:42:02.410248 139715572925888 deprecation_wrapper.py:119] From /home/jupyter/training-data-analyst/courses/machine_learning/deepdive/05_artandscience/house_prediction_module/trainer/model.py:41: The name tf.train.FtrlOptimizer is deprecated. Please use tf.compat.v1.train.FtrlOptimizer instead.\n\nI0809 20:42:02.410758 139715572925888 run_config.py:528] TF_CONFIG environment variable: {u'environment': u'cloud', u'cluster': {}, u'job': {u'args': [u'--batch_size=30', u'--learning_rate=0.02', u'--output_dir=house_trained', u'--job-dir', u'house_trained'], u'job_name': u'trainer.task'}, u'task': {}}\nI0809 20:42:02.411099 139715572925888 estimator.py:1790] Using default config.\nI0809 20:42:02.412035 139715572925888 estimator.py:209] Using config: {'_save_checkpoints_secs': 600, '_num_ps_replicas': 0, '_keep_checkpoint_max': 5, '_task_type': 'worker', '_global_id_in_cluster': 0, '_is_chief': True, '_cluster_spec': <tensorflow.python.training.server_lib.ClusterSpec object at 0x7f11d87aea50>, '_model_dir': 'house_trained/', '_protocol': None, '_save_checkpoints_steps': None, '_keep_checkpoint_every_n_hours': 10000, '_service': None, '_session_config': allow_soft_placement: true\ngraph_options {\n rewrite_options {\n meta_optimizer_iterations: ONE\n }\n}\n, '_tf_random_seed': None, '_save_summary_steps': 100, '_device_fn': None, '_experimental_distribute': None, '_num_worker_replicas': 1, '_task_id': 0, '_log_step_count_steps': 100, '_experimental_max_worker_delay_secs': None, '_evaluation_master': '', '_eval_distribute': None, '_train_distribute': None, '_master': ''}\nW0809 20:42:03.567886 139715572925888 lazy_loader.py:50] \nThe TensorFlow contrib module will not be included in TensorFlow 2.0.\nFor more information, please see:\n * https://github.com/tensorflow/community/blob/master/rfcs/20180907-contrib-sunset.md\n * https://github.com/tensorflow/addons\n * https://github.com/tensorflow/io (for I/O related ops)\nIf you depend on functionality not listed there, please file an issue.\n\nI0809 20:42:03.568871 139715572925888 estimator.py:209] Using config: {'_save_checkpoints_secs': 600, '_num_ps_replicas': 0, '_keep_checkpoint_max': 5, '_task_type': 'worker', '_global_id_in_cluster': 0, '_is_chief': True, '_cluster_spec': <tensorflow.python.training.server_lib.ClusterSpec object at 0x7f11d06b4110>, '_model_dir': 'house_trained/', '_protocol': None, '_save_checkpoints_steps': None, '_keep_checkpoint_every_n_hours': 10000, '_service': None, '_session_config': allow_soft_placement: true\ngraph_options {\n rewrite_options {\n meta_optimizer_iterations: ONE\n }\n}\n, '_tf_random_seed': None, '_save_summary_steps': 100, '_device_fn': None, '_experimental_distribute': None, '_num_worker_replicas': 1, '_task_id': 0, '_log_step_count_steps': 100, '_experimental_max_worker_delay_secs': None, '_evaluation_master': '', '_eval_distribute': None, '_train_distribute': None, '_master': ''}\nW0809 20:42:03.569215 139715572925888 deprecation_wrapper.py:119] From /home/jupyter/training-data-analyst/courses/machine_learning/deepdive/05_artandscience/house_prediction_module/trainer/model.py:20: The name tf.estimator.inputs is deprecated. Please use tf.compat.v1.estimator.inputs instead.\n\nW0809 20:42:03.569324 139715572925888 deprecation_wrapper.py:119] From /home/jupyter/training-data-analyst/courses/machine_learning/deepdive/05_artandscience/house_prediction_module/trainer/model.py:20: The name tf.estimator.inputs.pandas_input_fn is deprecated. Please use tf.compat.v1.estimator.inputs.pandas_input_fn instead.\n\nI0809 20:42:03.577970 139715572925888 estimator_training.py:186] Not using Distribute Coordinator.\nI0809 20:42:03.578327 139715572925888 training.py:612] Running training and evaluation locally (non-distributed).\nI0809 20:42:03.578629 139715572925888 training.py:700] Start train and evaluate loop. The evaluate will happen after every checkpoint. Checkpoint frequency is determined based on RunConfig arguments: save_checkpoints_steps None or save_checkpoints_secs 600.\nW0809 20:42:03.585417 139715572925888 deprecation.py:323] From /usr/local/lib/python2.7/dist-packages/tensorflow/python/training/training_util.py:236: initialized_value (from tensorflow.python.ops.variables) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse Variable.read_value. Variables in 2.X are initialized automatically both in eager and graph (inside tf.defun) contexts.\nW0809 20:42:03.600763 139715572925888 deprecation.py:323] From /usr/local/lib/python2.7/dist-packages/tensorflow_estimator/python/estimator/inputs/queues/feeding_queue_runner.py:62: __init__ (from tensorflow.python.training.queue_runner_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nTo construct input pipelines, use the `tf.data` module.\nW0809 20:42:03.601921 139715572925888 deprecation.py:323] From /usr/local/lib/python2.7/dist-packages/tensorflow_estimator/python/estimator/inputs/queues/feeding_functions.py:500: add_queue_runner (from tensorflow.python.training.queue_runner_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nTo construct input pipelines, use the `tf.data` module.\nI0809 20:42:03.610526 139715572925888 estimator.py:1145] Calling model_fn.\nI0809 20:42:03.610791 139715572925888 estimator.py:1145] Calling model_fn.\nW0809 20:42:03.937570 139715572925888 deprecation.py:323] From /usr/local/lib/python2.7/dist-packages/tensorflow_estimator/python/estimator/canned/linear.py:308: to_float (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `tf.cast` instead.\nI0809 20:42:04.041716 139715572925888 estimator.py:1147] Done calling model_fn.\nI0809 20:42:04.041951 139715572925888 estimator.py:1147] Done calling model_fn.\nI0809 20:42:04.042213 139715572925888 basic_session_run_hooks.py:541] Create CheckpointSaverHook.\nW0809 20:42:04.096678 139715572925888 deprecation.py:323] From /usr/local/lib/python2.7/dist-packages/tensorflow/python/ops/array_ops.py:1354: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\nI0809 20:42:04.321468 139715572925888 monitored_session.py:240] Graph was finalized.\n2019-08-09 20:42:04.321920: I tensorflow/core/platform/cpu_feature_guard.cc:145] This TensorFlow binary is optimized with Intel(R) MKL-DNN to use the following CPU instructions in performance critical operations: AVX2 FMA\nTo enable them in non-MKL-DNN operations, rebuild TensorFlow with the appropriate compiler flags.\n2019-08-09 20:42:04.331591: I tensorflow/core/platform/profile_utils/cpu_utils.cc:94] CPU Frequency: 2200000000 Hz\n2019-08-09 20:42:04.332354: I tensorflow/compiler/xla/service/service.cc:168] XLA service 0x5575845a21f0 executing computations on platform Host. Devices:\n2019-08-09 20:42:04.332417: I tensorflow/compiler/xla/service/service.cc:175] StreamExecutor device (0): <undefined>, <undefined>\n2019-08-09 20:42:04.332908: I tensorflow/core/common_runtime/process_util.cc:115] Creating new thread pool with default inter op setting: 2. Tune using inter_op_parallelism_threads for best performance.\n2019-08-09 20:42:04.355002: W tensorflow/compiler/jit/mark_for_compilation_pass.cc:1412] (One-time warning): Not using XLA:CPU for cluster because envvar TF_XLA_FLAGS=--tf_xla_cpu_global_jit was not set. If you want XLA:CPU, either set that envvar, or use experimental_jit_scope to enable XLA:CPU. To confirm that XLA is active, pass --vmodule=xla_compilation_cache=1 (as a proper command-line flag, not via TF_XLA_FLAGS) or set the envvar XLA_FLAGS=--xla_hlo_profile.\nI0809 20:42:04.382742 139715572925888 session_manager.py:500] Running local_init_op.\nI0809 20:42:04.388341 139715572925888 session_manager.py:502] Done running local_init_op.\nW0809 20:42:04.411591 139715572925888 deprecation.py:323] From /usr/local/lib/python2.7/dist-packages/tensorflow/python/training/monitored_session.py:875: start_queue_runners (from tensorflow.python.training.queue_runner_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nTo construct input pipelines, use the `tf.data` module.\nI0809 20:42:04.641765 139715572925888 basic_session_run_hooks.py:606] Saving checkpoints for 0 into house_trained/model.ckpt.\nI0809 20:42:04.843869 139715572925888 basic_session_run_hooks.py:262] loss = 215.66043, step = 1\nW0809 20:42:04.956371 139715572925888 basic_session_run_hooks.py:724] It seems that global step (tf.train.get_global_step) has not been increased. Current value (could be stable): 18 vs previous value: 18. You could increase the global step by passing tf.train.get_global_step() to Optimizer.apply_gradients or Optimizer.minimize.\nW0809 20:42:05.048196 139715572925888 basic_session_run_hooks.py:724] It seems that global step (tf.train.get_global_step) has not been increased. Current value (could be stable): 57 vs previous value: 57. You could increase the global step by passing tf.train.get_global_step() to Optimizer.apply_gradients or Optimizer.minimize.\nW0809 20:42:05.063069 139715572925888 basic_session_run_hooks.py:724] It seems that global step (tf.train.get_global_step) has not been increased. Current value (could be stable): 63 vs previous value: 63. You could increase the global step by passing tf.train.get_global_step() to Optimizer.apply_gradients or Optimizer.minimize.\nW0809 20:42:05.076215 139715572925888 basic_session_run_hooks.py:724] It seems that global step (tf.train.get_global_step) has not been increased. Current value (could be stable): 68 vs previous value: 68. You could increase the global step by passing tf.train.get_global_step() to Optimizer.apply_gradients or Optimizer.minimize.\nW0809 20:42:05.114201 139715572925888 basic_session_run_hooks.py:724] It seems that global step (tf.train.get_global_step) has not been increased. Current value (could be stable): 83 vs previous value: 83. You could increase the global step by passing tf.train.get_global_step() to Optimizer.apply_gradients or Optimizer.minimize.\nI0809 20:42:05.157850 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 317.933\nI0809 20:42:05.159033 139715572925888 basic_session_run_hooks.py:260] loss = 54.358315, step = 101 (0.315 sec)\nI0809 20:42:05.406924 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 401.551\nI0809 20:42:05.408078 139715572925888 basic_session_run_hooks.py:260] loss = 42.23906, step = 201 (0.249 sec)\nI0809 20:42:05.646493 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 417.343\nI0809 20:42:05.647548 139715572925888 basic_session_run_hooks.py:260] loss = 43.14472, step = 301 (0.239 sec)\nI0809 20:42:05.883550 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 421.896\nI0809 20:42:05.884644 139715572925888 basic_session_run_hooks.py:260] loss = 54.47378, step = 401 (0.237 sec)\nI0809 20:42:06.129745 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 406.174\nI0809 20:42:06.130978 139715572925888 basic_session_run_hooks.py:260] loss = 14.438426, step = 501 (0.246 sec)\nI0809 20:42:06.358213 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 437.652\nI0809 20:42:06.359368 139715572925888 basic_session_run_hooks.py:260] loss = 57.73707, step = 601 (0.228 sec)\nI0809 20:42:06.575443 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 460.34\nI0809 20:42:06.576554 139715572925888 basic_session_run_hooks.py:260] loss = 22.231636, step = 701 (0.217 sec)\nI0809 20:42:06.798619 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 448.171\nI0809 20:42:06.800406 139715572925888 basic_session_run_hooks.py:260] loss = 54.715797, step = 801 (0.224 sec)\nI0809 20:42:07.043385 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 408.513\nI0809 20:42:07.044590 139715572925888 basic_session_run_hooks.py:260] loss = 28.722849, step = 901 (0.244 sec)\nI0809 20:42:07.289911 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 405.596\nI0809 20:42:07.290906 139715572925888 basic_session_run_hooks.py:260] loss = 26.559034, step = 1001 (0.246 sec)\nI0809 20:42:07.529155 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 418.015\nI0809 20:42:07.530375 139715572925888 basic_session_run_hooks.py:260] loss = 1241.5792, step = 1101 (0.239 sec)\nI0809 20:42:07.781065 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 396.961\nI0809 20:42:07.782162 139715572925888 basic_session_run_hooks.py:260] loss = 29.28805, step = 1201 (0.252 sec)\nI0809 20:42:08.019126 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 420.016\nI0809 20:42:08.020097 139715572925888 basic_session_run_hooks.py:260] loss = 37.746925, step = 1301 (0.238 sec)\nI0809 20:42:08.244482 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 443.754\nI0809 20:42:08.247842 139715572925888 basic_session_run_hooks.py:260] loss = 24.188057, step = 1401 (0.228 sec)\nI0809 20:42:08.498735 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 393.352\nI0809 20:42:08.499910 139715572925888 basic_session_run_hooks.py:260] loss = 60.33488, step = 1501 (0.252 sec)\nI0809 20:42:08.721335 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 449.228\nI0809 20:42:08.722378 139715572925888 basic_session_run_hooks.py:260] loss = 21.831383, step = 1601 (0.222 sec)\nI0809 20:42:08.941145 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 455.438\nI0809 20:42:08.942961 139715572925888 basic_session_run_hooks.py:260] loss = 60.54083, step = 1701 (0.221 sec)\nI0809 20:42:09.173693 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 429.533\nI0809 20:42:09.174632 139715572925888 basic_session_run_hooks.py:260] loss = 34.44056, step = 1801 (0.232 sec)\nI0809 20:42:09.421431 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 403.67\nI0809 20:42:09.422734 139715572925888 basic_session_run_hooks.py:260] loss = 16.504276, step = 1901 (0.248 sec)\nI0809 20:42:09.635639 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 466.79\nI0809 20:42:09.636759 139715572925888 basic_session_run_hooks.py:260] loss = 62.338196, step = 2001 (0.214 sec)\nI0809 20:42:09.856868 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 452.031\nI0809 20:42:09.857850 139715572925888 basic_session_run_hooks.py:260] loss = 34.891525, step = 2101 (0.221 sec)\nI0809 20:42:10.070991 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 468.876\nI0809 20:42:10.072278 139715572925888 basic_session_run_hooks.py:260] loss = 36.803764, step = 2201 (0.214 sec)\nI0809 20:42:10.292645 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 449.408\nI0809 20:42:10.293653 139715572925888 basic_session_run_hooks.py:260] loss = 19.011322, step = 2301 (0.221 sec)\nI0809 20:42:10.504937 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 471.143\nI0809 20:42:10.507251 139715572925888 basic_session_run_hooks.py:260] loss = 50.321453, step = 2401 (0.214 sec)\nI0809 20:42:10.739464 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 426.397\nI0809 20:42:10.740521 139715572925888 basic_session_run_hooks.py:260] loss = 84.55872, step = 2501 (0.233 sec)\nI0809 20:42:10.979623 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 416.332\nI0809 20:42:10.980529 139715572925888 basic_session_run_hooks.py:260] loss = 50.548977, step = 2601 (0.240 sec)\nI0809 20:42:11.199657 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 454.505\nI0809 20:42:11.200752 139715572925888 basic_session_run_hooks.py:260] loss = 41.289875, step = 2701 (0.220 sec)\nI0809 20:42:11.416954 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 460.337\nI0809 20:42:11.418008 139715572925888 basic_session_run_hooks.py:260] loss = 15.092587, step = 2801 (0.217 sec)\nI0809 20:42:11.651057 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 427.035\nI0809 20:42:11.652159 139715572925888 basic_session_run_hooks.py:260] loss = 66.30819, step = 2901 (0.234 sec)\nI0809 20:42:11.894701 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 410.46\nI0809 20:42:11.895662 139715572925888 basic_session_run_hooks.py:260] loss = 32.576336, step = 3001 (0.244 sec)\nI0809 20:42:12.132533 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 420.412\nI0809 20:42:12.133589 139715572925888 basic_session_run_hooks.py:260] loss = 31.308903, step = 3101 (0.238 sec)\nI0809 20:42:12.339224 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 483.828\nI0809 20:42:12.340104 139715572925888 basic_session_run_hooks.py:260] loss = 24.115883, step = 3201 (0.207 sec)\nI0809 20:42:12.568285 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 436.719\nI0809 20:42:12.569576 139715572925888 basic_session_run_hooks.py:260] loss = 20.761528, step = 3301 (0.229 sec)\nI0809 20:42:12.798980 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 433.377\nI0809 20:42:12.800106 139715572925888 basic_session_run_hooks.py:260] loss = 31.681124, step = 3401 (0.231 sec)\nI0809 20:42:13.030879 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 431.155\nI0809 20:42:13.031781 139715572925888 basic_session_run_hooks.py:260] loss = 22.891483, step = 3501 (0.232 sec)\nI0809 20:42:13.280486 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 400.651\nI0809 20:42:13.281569 139715572925888 basic_session_run_hooks.py:260] loss = 41.06246, step = 3601 (0.250 sec)\nI0809 20:42:13.519926 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 417.697\nI0809 20:42:13.521043 139715572925888 basic_session_run_hooks.py:260] loss = 21.739857, step = 3701 (0.239 sec)\nI0809 20:42:13.749242 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 436.021\nI0809 20:42:13.750402 139715572925888 basic_session_run_hooks.py:260] loss = 127.92703, step = 3801 (0.229 sec)\nI0809 20:42:13.978998 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 435.309\nI0809 20:42:13.980202 139715572925888 basic_session_run_hooks.py:260] loss = 14.991419, step = 3901 (0.230 sec)\nI0809 20:42:14.206845 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 438.824\nI0809 20:42:14.207771 139715572925888 basic_session_run_hooks.py:260] loss = 36.550327, step = 4001 (0.228 sec)\nI0809 20:42:14.428431 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 451.738\nI0809 20:42:14.429389 139715572925888 basic_session_run_hooks.py:260] loss = 39.62497, step = 4101 (0.222 sec)\nI0809 20:42:14.659174 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 432.968\nI0809 20:42:14.660362 139715572925888 basic_session_run_hooks.py:260] loss = 32.269123, step = 4201 (0.231 sec)\nI0809 20:42:14.895462 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 423.278\nI0809 20:42:14.897689 139715572925888 basic_session_run_hooks.py:260] loss = 86.88386, step = 4301 (0.237 sec)\nI0809 20:42:15.108449 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 469.492\nI0809 20:42:15.109358 139715572925888 basic_session_run_hooks.py:260] loss = 39.07176, step = 4401 (0.212 sec)\nI0809 20:42:15.340329 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 431.263\nI0809 20:42:15.341247 139715572925888 basic_session_run_hooks.py:260] loss = 47.81019, step = 4501 (0.232 sec)\nI0809 20:42:15.562230 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 450.641\nI0809 20:42:15.563405 139715572925888 basic_session_run_hooks.py:260] loss = 31.643383, step = 4601 (0.222 sec)\nI0809 20:42:15.806593 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 409.209\nI0809 20:42:15.807668 139715572925888 basic_session_run_hooks.py:260] loss = 38.677456, step = 4701 (0.244 sec)\nI0809 20:42:16.040714 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 427.341\nI0809 20:42:16.042851 139715572925888 basic_session_run_hooks.py:260] loss = 20.32229, step = 4801 (0.235 sec)\nI0809 20:42:16.278851 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 419.746\nI0809 20:42:16.280090 139715572925888 basic_session_run_hooks.py:260] loss = 70.01985, step = 4901 (0.237 sec)\nI0809 20:42:16.520617 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 413.597\nI0809 20:42:16.521884 139715572925888 basic_session_run_hooks.py:260] loss = 23.471846, step = 5001 (0.242 sec)\nI0809 20:42:16.759655 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 418.356\nI0809 20:42:16.760668 139715572925888 basic_session_run_hooks.py:260] loss = 19.1656, step = 5101 (0.239 sec)\nI0809 20:42:16.991517 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 431.241\nI0809 20:42:16.992455 139715572925888 basic_session_run_hooks.py:260] loss = 37.272438, step = 5201 (0.232 sec)\nI0809 20:42:17.241883 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 399.484\nI0809 20:42:17.243081 139715572925888 basic_session_run_hooks.py:260] loss = 47.863422, step = 5301 (0.251 sec)\nI0809 20:42:17.475157 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 428.721\nI0809 20:42:17.477113 139715572925888 basic_session_run_hooks.py:260] loss = 20.762028, step = 5401 (0.234 sec)\nI0809 20:42:17.706330 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 432.509\nI0809 20:42:17.707281 139715572925888 basic_session_run_hooks.py:260] loss = 31.892517, step = 5501 (0.230 sec)\nI0809 20:42:17.922365 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 462.862\nI0809 20:42:17.923373 139715572925888 basic_session_run_hooks.py:260] loss = 49.485367, step = 5601 (0.216 sec)\nI0809 20:42:18.147011 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 445.177\nI0809 20:42:18.148031 139715572925888 basic_session_run_hooks.py:260] loss = 21.976042, step = 5701 (0.225 sec)\nI0809 20:42:18.384932 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 420.274\nI0809 20:42:18.386049 139715572925888 basic_session_run_hooks.py:260] loss = 47.708855, step = 5801 (0.238 sec)\nI0809 20:42:18.605120 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 454.131\nI0809 20:42:18.606066 139715572925888 basic_session_run_hooks.py:260] loss = 38.32365, step = 5901 (0.220 sec)\nI0809 20:42:18.849222 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 409.718\nI0809 20:42:18.850502 139715572925888 basic_session_run_hooks.py:260] loss = 19.839962, step = 6001 (0.244 sec)\nI0809 20:42:19.075581 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 441.809\nI0809 20:42:19.076723 139715572925888 basic_session_run_hooks.py:260] loss = 35.977077, step = 6101 (0.226 sec)\nI0809 20:42:19.309335 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 427.707\nI0809 20:42:19.310457 139715572925888 basic_session_run_hooks.py:260] loss = 40.517097, step = 6201 (0.234 sec)\nI0809 20:42:19.544992 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 424.403\nI0809 20:42:19.546072 139715572925888 basic_session_run_hooks.py:260] loss = 28.959423, step = 6301 (0.236 sec)\nI0809 20:42:19.782182 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 421.605\nI0809 20:42:19.783279 139715572925888 basic_session_run_hooks.py:260] loss = 25.088074, step = 6401 (0.237 sec)\nI0809 20:42:20.022277 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 416.459\nI0809 20:42:20.023374 139715572925888 basic_session_run_hooks.py:260] loss = 51.164604, step = 6501 (0.240 sec)\nI0809 20:42:20.248764 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 441.532\nI0809 20:42:20.249742 139715572925888 basic_session_run_hooks.py:260] loss = 23.999224, step = 6601 (0.226 sec)\nI0809 20:42:20.469518 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 453.042\nI0809 20:42:20.470803 139715572925888 basic_session_run_hooks.py:260] loss = 103.26223, step = 6701 (0.221 sec)\nI0809 20:42:20.702929 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 428.391\nI0809 20:42:20.703821 139715572925888 basic_session_run_hooks.py:260] loss = 44.0259, step = 6801 (0.233 sec)\nI0809 20:42:20.949997 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 404.778\nI0809 20:42:20.951296 139715572925888 basic_session_run_hooks.py:260] loss = 19.171732, step = 6901 (0.247 sec)\nI0809 20:42:21.216790 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 374.786\nI0809 20:42:21.217852 139715572925888 basic_session_run_hooks.py:260] loss = 122.0697, step = 7001 (0.267 sec)\nI0809 20:42:21.435852 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 456.642\nI0809 20:42:21.437046 139715572925888 basic_session_run_hooks.py:260] loss = 33.82604, step = 7101 (0.219 sec)\nI0809 20:42:21.669198 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 428.495\nI0809 20:42:21.670361 139715572925888 basic_session_run_hooks.py:260] loss = 24.399328, step = 7201 (0.233 sec)\nI0809 20:42:21.912559 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 410.811\nI0809 20:42:21.913444 139715572925888 basic_session_run_hooks.py:260] loss = 34.948128, step = 7301 (0.243 sec)\nI0809 20:42:22.130326 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 459.228\nI0809 20:42:22.131231 139715572925888 basic_session_run_hooks.py:260] loss = 72.025986, step = 7401 (0.218 sec)\nI0809 20:42:22.342922 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 470.332\nI0809 20:42:22.343796 139715572925888 basic_session_run_hooks.py:260] loss = 23.518652, step = 7501 (0.213 sec)\nI0809 20:42:22.550642 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 481.533\nI0809 20:42:22.551698 139715572925888 basic_session_run_hooks.py:260] loss = 47.13624, step = 7601 (0.208 sec)\nI0809 20:42:22.765336 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 465.694\nI0809 20:42:22.766264 139715572925888 basic_session_run_hooks.py:260] loss = 29.63544, step = 7701 (0.215 sec)\nI0809 20:42:22.983797 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 457.74\nI0809 20:42:22.984637 139715572925888 basic_session_run_hooks.py:260] loss = 14.424541, step = 7801 (0.218 sec)\nI0809 20:42:23.160651 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 565.544\nI0809 20:42:23.162517 139715572925888 basic_session_run_hooks.py:260] loss = 68.20079, step = 7901 (0.178 sec)\nI0809 20:42:23.371675 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 473.925\nI0809 20:42:23.372550 139715572925888 basic_session_run_hooks.py:260] loss = 43.285156, step = 8001 (0.210 sec)\nI0809 20:42:23.575895 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 489.574\nI0809 20:42:23.576782 139715572925888 basic_session_run_hooks.py:260] loss = 29.779673, step = 8101 (0.204 sec)\nI0809 20:42:23.760229 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 542.529\nI0809 20:42:23.761007 139715572925888 basic_session_run_hooks.py:260] loss = 18.886053, step = 8201 (0.184 sec)\nI0809 20:42:23.954575 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 514.57\nI0809 20:42:23.955420 139715572925888 basic_session_run_hooks.py:260] loss = 61.837727, step = 8301 (0.194 sec)\nI0809 20:42:24.172730 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 458.442\nI0809 20:42:24.175014 139715572925888 basic_session_run_hooks.py:260] loss = 35.335217, step = 8401 (0.220 sec)\nI0809 20:42:24.363615 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 523.728\nI0809 20:42:24.364455 139715572925888 basic_session_run_hooks.py:260] loss = 73.91724, step = 8501 (0.189 sec)\nI0809 20:42:24.545725 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 549.106\nI0809 20:42:24.546555 139715572925888 basic_session_run_hooks.py:260] loss = 32.35276, step = 8601 (0.182 sec)\nI0809 20:42:24.728656 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 546.699\nI0809 20:42:24.729615 139715572925888 basic_session_run_hooks.py:260] loss = 9.280526, step = 8701 (0.183 sec)\nI0809 20:42:24.917541 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 529.392\nI0809 20:42:24.918524 139715572925888 basic_session_run_hooks.py:260] loss = 52.10976, step = 8801 (0.189 sec)\nI0809 20:42:25.101804 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 542.696\nI0809 20:42:25.102653 139715572925888 basic_session_run_hooks.py:260] loss = 31.689117, step = 8901 (0.184 sec)\nI0809 20:42:25.308911 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 482.814\nI0809 20:42:25.309767 139715572925888 basic_session_run_hooks.py:260] loss = 53.8692, step = 9001 (0.207 sec)\nI0809 20:42:25.512624 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 490.993\nI0809 20:42:25.514770 139715572925888 basic_session_run_hooks.py:260] loss = 29.0009, step = 9101 (0.205 sec)\nI0809 20:42:25.703310 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 524.346\nI0809 20:42:25.704185 139715572925888 basic_session_run_hooks.py:260] loss = 27.948374, step = 9201 (0.189 sec)\nI0809 20:42:25.884633 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 551.514\nI0809 20:42:25.885483 139715572925888 basic_session_run_hooks.py:260] loss = 34.51767, step = 9301 (0.181 sec)\nI0809 20:42:26.088884 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 489.596\nI0809 20:42:26.089658 139715572925888 basic_session_run_hooks.py:260] loss = 40.559605, step = 9401 (0.204 sec)\nI0809 20:42:26.288553 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 500.786\nI0809 20:42:26.289357 139715572925888 basic_session_run_hooks.py:260] loss = 56.587803, step = 9501 (0.200 sec)\nI0809 20:42:26.471863 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 545.548\nI0809 20:42:26.472719 139715572925888 basic_session_run_hooks.py:260] loss = 30.86315, step = 9601 (0.183 sec)\nI0809 20:42:26.654179 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 548.537\nI0809 20:42:26.654956 139715572925888 basic_session_run_hooks.py:260] loss = 42.84465, step = 9701 (0.182 sec)\nI0809 20:42:26.836205 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 549.348\nI0809 20:42:26.836946 139715572925888 basic_session_run_hooks.py:260] loss = 32.10369, step = 9801 (0.182 sec)\nI0809 20:42:27.029071 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 518.444\nI0809 20:42:27.029989 139715572925888 basic_session_run_hooks.py:260] loss = 50.195885, step = 9901 (0.193 sec)\nI0809 20:42:27.224503 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 511.713\nI0809 20:42:27.225347 139715572925888 basic_session_run_hooks.py:260] loss = 21.120586, step = 10001 (0.195 sec)\nI0809 20:42:27.402610 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 561.508\nI0809 20:42:27.403367 139715572925888 basic_session_run_hooks.py:260] loss = 48.493736, step = 10101 (0.178 sec)\nI0809 20:42:27.594835 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 520.208\nI0809 20:42:27.595704 139715572925888 basic_session_run_hooks.py:260] loss = 27.813099, step = 10201 (0.192 sec)\nI0809 20:42:27.796318 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 496.307\nI0809 20:42:27.797118 139715572925888 basic_session_run_hooks.py:260] loss = 27.684166, step = 10301 (0.201 sec)\nI0809 20:42:28.001615 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 487.151\nI0809 20:42:28.002585 139715572925888 basic_session_run_hooks.py:260] loss = 41.948124, step = 10401 (0.205 sec)\nI0809 20:42:28.189969 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 530.828\nI0809 20:42:28.190793 139715572925888 basic_session_run_hooks.py:260] loss = 19.98607, step = 10501 (0.188 sec)\nI0809 20:42:28.365161 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 570.867\nI0809 20:42:28.366378 139715572925888 basic_session_run_hooks.py:260] loss = 87.41141, step = 10601 (0.176 sec)\nI0809 20:42:28.568749 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 491.147\nI0809 20:42:28.569617 139715572925888 basic_session_run_hooks.py:260] loss = 27.779526, step = 10701 (0.203 sec)\nI0809 20:42:28.772715 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 490.268\nI0809 20:42:28.773514 139715572925888 basic_session_run_hooks.py:260] loss = 32.40663, step = 10801 (0.204 sec)\nI0809 20:42:28.960273 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 533.254\nI0809 20:42:28.961036 139715572925888 basic_session_run_hooks.py:260] loss = 8.738708, step = 10901 (0.188 sec)\nI0809 20:42:29.170430 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 475.769\nI0809 20:42:29.171308 139715572925888 basic_session_run_hooks.py:260] loss = 17.224623, step = 11001 (0.210 sec)\nI0809 20:42:29.378458 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 480.693\nI0809 20:42:29.379267 139715572925888 basic_session_run_hooks.py:260] loss = 129.82169, step = 11101 (0.208 sec)\nI0809 20:42:29.581667 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 492.16\nI0809 20:42:29.582468 139715572925888 basic_session_run_hooks.py:260] loss = 59.290813, step = 11201 (0.203 sec)\nI0809 20:42:29.772501 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 524.053\nI0809 20:42:29.773473 139715572925888 basic_session_run_hooks.py:260] loss = 48.67883, step = 11301 (0.191 sec)\nI0809 20:42:29.952811 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 554.551\nI0809 20:42:29.953942 139715572925888 basic_session_run_hooks.py:260] loss = 31.161797, step = 11401 (0.180 sec)\nI0809 20:42:30.128308 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 569.762\nI0809 20:42:30.129107 139715572925888 basic_session_run_hooks.py:260] loss = 85.07748, step = 11501 (0.175 sec)\nI0809 20:42:30.307543 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 557.967\nI0809 20:42:30.308506 139715572925888 basic_session_run_hooks.py:260] loss = 37.616295, step = 11601 (0.179 sec)\nI0809 20:42:30.481650 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 574.422\nI0809 20:42:30.482454 139715572925888 basic_session_run_hooks.py:260] loss = 73.01372, step = 11701 (0.174 sec)\nI0809 20:42:30.664022 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 548.27\nI0809 20:42:30.665050 139715572925888 basic_session_run_hooks.py:260] loss = 35.499306, step = 11801 (0.183 sec)\nI0809 20:42:30.870676 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 483.863\nI0809 20:42:30.871476 139715572925888 basic_session_run_hooks.py:260] loss = 18.893257, step = 11901 (0.206 sec)\nI0809 20:42:31.054171 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 544.962\nI0809 20:42:31.054955 139715572925888 basic_session_run_hooks.py:260] loss = 70.91739, step = 12001 (0.183 sec)\nI0809 20:42:31.227020 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 578.58\nI0809 20:42:31.227952 139715572925888 basic_session_run_hooks.py:260] loss = 47.56703, step = 12101 (0.173 sec)\nI0809 20:42:31.437911 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 474.165\nI0809 20:42:31.438690 139715572925888 basic_session_run_hooks.py:260] loss = 21.411425, step = 12201 (0.211 sec)\nI0809 20:42:31.643642 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 486.081\nI0809 20:42:31.644399 139715572925888 basic_session_run_hooks.py:260] loss = 14.617336, step = 12301 (0.206 sec)\nI0809 20:42:31.819405 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 568.928\nI0809 20:42:31.820190 139715572925888 basic_session_run_hooks.py:260] loss = 64.563515, step = 12401 (0.176 sec)\nI0809 20:42:31.993980 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 572.797\nI0809 20:42:31.994754 139715572925888 basic_session_run_hooks.py:260] loss = 30.201702, step = 12501 (0.175 sec)\nI0809 20:42:32.177431 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 545.191\nI0809 20:42:32.178247 139715572925888 basic_session_run_hooks.py:260] loss = 62.845848, step = 12601 (0.183 sec)\nI0809 20:42:32.356033 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 559.838\nI0809 20:42:32.356898 139715572925888 basic_session_run_hooks.py:260] loss = 28.99841, step = 12701 (0.179 sec)\nI0809 20:42:32.530317 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 573.773\nI0809 20:42:32.531068 139715572925888 basic_session_run_hooks.py:260] loss = 47.354977, step = 12801 (0.174 sec)\nI0809 20:42:32.706470 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 567.717\nI0809 20:42:32.707315 139715572925888 basic_session_run_hooks.py:260] loss = 61.95388, step = 12901 (0.176 sec)\nI0809 20:42:32.881021 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 572.899\nI0809 20:42:32.881838 139715572925888 basic_session_run_hooks.py:260] loss = 38.48935, step = 13001 (0.175 sec)\nI0809 20:42:33.056893 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 568.676\nI0809 20:42:33.057935 139715572925888 basic_session_run_hooks.py:260] loss = 31.168634, step = 13101 (0.176 sec)\nI0809 20:42:33.240495 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 544.534\nI0809 20:42:33.241319 139715572925888 basic_session_run_hooks.py:260] loss = 28.266655, step = 13201 (0.183 sec)\nI0809 20:42:33.449738 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 477.941\nI0809 20:42:33.450530 139715572925888 basic_session_run_hooks.py:260] loss = 81.25282, step = 13301 (0.209 sec)\nI0809 20:42:33.640973 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 522.876\nI0809 20:42:33.641772 139715572925888 basic_session_run_hooks.py:260] loss = 26.1285, step = 13401 (0.191 sec)\nI0809 20:42:33.839467 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 503.849\nI0809 20:42:33.840281 139715572925888 basic_session_run_hooks.py:260] loss = 44.024464, step = 13501 (0.199 sec)\nI0809 20:42:34.032309 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 518.535\nI0809 20:42:34.033133 139715572925888 basic_session_run_hooks.py:260] loss = 31.122879, step = 13601 (0.193 sec)\nI0809 20:42:34.240973 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 479.249\nI0809 20:42:34.241849 139715572925888 basic_session_run_hooks.py:260] loss = 10.125617, step = 13701 (0.209 sec)\nI0809 20:42:34.434063 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 517.866\nI0809 20:42:34.434878 139715572925888 basic_session_run_hooks.py:260] loss = 70.14729, step = 13801 (0.193 sec)\nI0809 20:42:34.621891 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 532.453\nI0809 20:42:34.622862 139715572925888 basic_session_run_hooks.py:260] loss = 27.217846, step = 13901 (0.188 sec)\nI0809 20:42:34.797688 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 568.896\nI0809 20:42:34.799663 139715572925888 basic_session_run_hooks.py:260] loss = 35.97609, step = 14001 (0.177 sec)\nI0809 20:42:34.979423 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 550.543\nI0809 20:42:34.980413 139715572925888 basic_session_run_hooks.py:260] loss = 26.12788, step = 14101 (0.181 sec)\nI0809 20:42:35.180140 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 497.973\nI0809 20:42:35.180926 139715572925888 basic_session_run_hooks.py:260] loss = 64.12424, step = 14201 (0.201 sec)\nI0809 20:42:35.361011 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 552.826\nI0809 20:42:35.362108 139715572925888 basic_session_run_hooks.py:260] loss = 32.04691, step = 14301 (0.181 sec)\nI0809 20:42:35.539427 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 560.453\nI0809 20:42:35.540393 139715572925888 basic_session_run_hooks.py:260] loss = 52.47918, step = 14401 (0.178 sec)\nI0809 20:42:35.743329 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 490.417\nI0809 20:42:35.744175 139715572925888 basic_session_run_hooks.py:260] loss = 24.953758, step = 14501 (0.204 sec)\nI0809 20:42:35.940030 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 508.443\nI0809 20:42:35.940902 139715572925888 basic_session_run_hooks.py:260] loss = 34.942688, step = 14601 (0.197 sec)\nI0809 20:42:36.117806 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 562.461\nI0809 20:42:36.118593 139715572925888 basic_session_run_hooks.py:260] loss = 55.465492, step = 14701 (0.178 sec)\nI0809 20:42:36.316770 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 502.589\nI0809 20:42:36.317580 139715572925888 basic_session_run_hooks.py:260] loss = 57.6858, step = 14801 (0.199 sec)\nI0809 20:42:36.524956 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 480.443\nI0809 20:42:36.527163 139715572925888 basic_session_run_hooks.py:260] loss = 35.631615, step = 14901 (0.210 sec)\nI0809 20:42:36.708018 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 546.17\nI0809 20:42:36.708904 139715572925888 basic_session_run_hooks.py:260] loss = 17.542585, step = 15001 (0.182 sec)\nI0809 20:42:36.890199 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 548.863\nI0809 20:42:36.890944 139715572925888 basic_session_run_hooks.py:260] loss = 53.94641, step = 15101 (0.182 sec)\nI0809 20:42:37.079205 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 529.118\nI0809 20:42:37.080248 139715572925888 basic_session_run_hooks.py:260] loss = 33.645924, step = 15201 (0.189 sec)\nI0809 20:42:37.256110 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 565.269\nI0809 20:42:37.256959 139715572925888 basic_session_run_hooks.py:260] loss = 45.093517, step = 15301 (0.177 sec)\nI0809 20:42:37.439413 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 545.554\nI0809 20:42:37.440299 139715572925888 basic_session_run_hooks.py:260] loss = 34.845398, step = 15401 (0.183 sec)\nI0809 20:42:37.642906 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 491.379\nI0809 20:42:37.643713 139715572925888 basic_session_run_hooks.py:260] loss = 10.090223, step = 15501 (0.203 sec)\nI0809 20:42:37.843957 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 497.42\nI0809 20:42:37.844818 139715572925888 basic_session_run_hooks.py:260] loss = 51.057957, step = 15601 (0.201 sec)\nI0809 20:42:38.021238 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 564.099\nI0809 20:42:38.022386 139715572925888 basic_session_run_hooks.py:260] loss = 37.113983, step = 15701 (0.178 sec)\nI0809 20:42:38.198247 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 564.892\nI0809 20:42:38.198960 139715572925888 basic_session_run_hooks.py:260] loss = 49.76435, step = 15801 (0.177 sec)\nI0809 20:42:38.373404 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 570.949\nI0809 20:42:38.374464 139715572925888 basic_session_run_hooks.py:260] loss = 16.172215, step = 15901 (0.176 sec)\nI0809 20:42:38.549565 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 567.679\nI0809 20:42:38.550405 139715572925888 basic_session_run_hooks.py:260] loss = 27.929382, step = 16001 (0.176 sec)\nI0809 20:42:38.727065 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 563.431\nI0809 20:42:38.727866 139715572925888 basic_session_run_hooks.py:260] loss = 39.391487, step = 16101 (0.177 sec)\nI0809 20:42:38.903073 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 568.05\nI0809 20:42:38.903871 139715572925888 basic_session_run_hooks.py:260] loss = 87.43811, step = 16201 (0.176 sec)\nI0809 20:42:39.086318 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 545.75\nI0809 20:42:39.087182 139715572925888 basic_session_run_hooks.py:260] loss = 29.880781, step = 16301 (0.183 sec)\nI0809 20:42:39.270591 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 542.682\nI0809 20:42:39.271349 139715572925888 basic_session_run_hooks.py:260] loss = 15.598202, step = 16401 (0.184 sec)\nI0809 20:42:39.459044 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 531.031\nI0809 20:42:39.459912 139715572925888 basic_session_run_hooks.py:260] loss = 50.94569, step = 16501 (0.189 sec)\nI0809 20:42:39.656713 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 505.533\nI0809 20:42:39.657488 139715572925888 basic_session_run_hooks.py:260] loss = 44.291523, step = 16601 (0.198 sec)\nI0809 20:42:39.839131 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 548.188\nI0809 20:42:39.839967 139715572925888 basic_session_run_hooks.py:260] loss = 86.52313, step = 16701 (0.182 sec)\nI0809 20:42:40.027645 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 530.481\nI0809 20:42:40.028501 139715572925888 basic_session_run_hooks.py:260] loss = 18.461927, step = 16801 (0.189 sec)\nI0809 20:42:40.249556 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 450.617\nI0809 20:42:40.250471 139715572925888 basic_session_run_hooks.py:260] loss = 36.88288, step = 16901 (0.222 sec)\nI0809 20:42:40.453813 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 489.634\nI0809 20:42:40.454663 139715572925888 basic_session_run_hooks.py:260] loss = 45.47953, step = 17001 (0.204 sec)\nI0809 20:42:40.655236 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 496.458\nI0809 20:42:40.656106 139715572925888 basic_session_run_hooks.py:260] loss = 18.319965, step = 17101 (0.201 sec)\nI0809 20:42:40.838766 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 544.849\nI0809 20:42:40.839612 139715572925888 basic_session_run_hooks.py:260] loss = 23.532417, step = 17201 (0.184 sec)\nI0809 20:42:41.045156 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 484.466\nI0809 20:42:41.045936 139715572925888 basic_session_run_hooks.py:260] loss = 21.925976, step = 17301 (0.206 sec)\nI0809 20:42:41.243024 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 505.398\nI0809 20:42:41.243818 139715572925888 basic_session_run_hooks.py:260] loss = 72.24338, step = 17401 (0.198 sec)\nI0809 20:42:41.429198 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 537.213\nI0809 20:42:41.429965 139715572925888 basic_session_run_hooks.py:260] loss = 23.276207, step = 17501 (0.186 sec)\nI0809 20:42:41.618097 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 529.325\nI0809 20:42:41.618875 139715572925888 basic_session_run_hooks.py:260] loss = 55.87291, step = 17601 (0.189 sec)\nI0809 20:42:41.810945 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 518.596\nI0809 20:42:41.811880 139715572925888 basic_session_run_hooks.py:260] loss = 28.344604, step = 17701 (0.193 sec)\nI0809 20:42:41.988164 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 564.341\nI0809 20:42:41.989120 139715572925888 basic_session_run_hooks.py:260] loss = 15.306099, step = 17801 (0.177 sec)\nI0809 20:42:42.194932 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 483.51\nI0809 20:42:42.195729 139715572925888 basic_session_run_hooks.py:260] loss = 50.228928, step = 17901 (0.207 sec)\nI0809 20:42:42.380472 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 539.031\nI0809 20:42:42.381618 139715572925888 basic_session_run_hooks.py:260] loss = 34.677082, step = 18001 (0.186 sec)\nI0809 20:42:42.559516 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 558.791\nI0809 20:42:42.560199 139715572925888 basic_session_run_hooks.py:260] loss = 34.266094, step = 18101 (0.179 sec)\nI0809 20:42:42.734060 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 572.586\nI0809 20:42:42.734889 139715572925888 basic_session_run_hooks.py:260] loss = 20.596783, step = 18201 (0.175 sec)\nI0809 20:42:42.908487 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 573.385\nI0809 20:42:42.909456 139715572925888 basic_session_run_hooks.py:260] loss = 95.104004, step = 18301 (0.175 sec)\nI0809 20:42:43.088056 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 556.827\nI0809 20:42:43.088960 139715572925888 basic_session_run_hooks.py:260] loss = 47.536537, step = 18401 (0.179 sec)\nI0809 20:42:43.290330 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 494.355\nI0809 20:42:43.291104 139715572925888 basic_session_run_hooks.py:260] loss = 63.25869, step = 18501 (0.202 sec)\nI0809 20:42:43.465354 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 571.38\nI0809 20:42:43.466170 139715572925888 basic_session_run_hooks.py:260] loss = 49.56587, step = 18601 (0.175 sec)\nI0809 20:42:43.659379 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 515.4\nI0809 20:42:43.660370 139715572925888 basic_session_run_hooks.py:260] loss = 19.38341, step = 18701 (0.194 sec)\nI0809 20:42:43.836994 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 563.047\nI0809 20:42:43.837881 139715572925888 basic_session_run_hooks.py:260] loss = 69.12962, step = 18801 (0.178 sec)\nI0809 20:42:44.014785 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 562.465\nI0809 20:42:44.015649 139715572925888 basic_session_run_hooks.py:260] loss = 43.236244, step = 18901 (0.178 sec)\nI0809 20:42:44.205502 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 524.304\nI0809 20:42:44.206520 139715572925888 basic_session_run_hooks.py:260] loss = 26.287243, step = 19001 (0.191 sec)\nI0809 20:42:44.390386 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 540.879\nI0809 20:42:44.391190 139715572925888 basic_session_run_hooks.py:260] loss = 13.419331, step = 19101 (0.185 sec)\nI0809 20:42:44.570664 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 554.708\nI0809 20:42:44.571482 139715572925888 basic_session_run_hooks.py:260] loss = 67.351204, step = 19201 (0.180 sec)\nI0809 20:42:44.752331 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 550.461\nI0809 20:42:44.753911 139715572925888 basic_session_run_hooks.py:260] loss = 22.54165, step = 19301 (0.182 sec)\nI0809 20:42:44.931735 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 557.407\nI0809 20:42:44.932554 139715572925888 basic_session_run_hooks.py:260] loss = 66.86839, step = 19401 (0.179 sec)\nI0809 20:42:45.108572 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 565.461\nI0809 20:42:45.109344 139715572925888 basic_session_run_hooks.py:260] loss = 24.244747, step = 19501 (0.177 sec)\nI0809 20:42:45.298085 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 527.68\nI0809 20:42:45.298973 139715572925888 basic_session_run_hooks.py:260] loss = 13.969262, step = 19601 (0.190 sec)\nI0809 20:42:45.492240 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 515.126\nI0809 20:42:45.493179 139715572925888 basic_session_run_hooks.py:260] loss = 76.86172, step = 19701 (0.194 sec)\nI0809 20:42:45.672214 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 555.661\nI0809 20:42:45.673048 139715572925888 basic_session_run_hooks.py:260] loss = 36.777737, step = 19801 (0.180 sec)\nI0809 20:42:45.850156 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 561.93\nI0809 20:42:45.851198 139715572925888 basic_session_run_hooks.py:260] loss = 26.178139, step = 19901 (0.178 sec)\nI0809 20:42:46.027370 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 564.242\nI0809 20:42:46.028316 139715572925888 basic_session_run_hooks.py:260] loss = 23.499826, step = 20001 (0.177 sec)\nI0809 20:42:46.214497 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 534.548\nI0809 20:42:46.216654 139715572925888 basic_session_run_hooks.py:260] loss = 50.725372, step = 20101 (0.188 sec)\nI0809 20:42:46.422430 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 480.834\nI0809 20:42:46.423405 139715572925888 basic_session_run_hooks.py:260] loss = 44.494545, step = 20201 (0.207 sec)\nI0809 20:42:46.599042 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 566.524\nI0809 20:42:46.599921 139715572925888 basic_session_run_hooks.py:260] loss = 33.04119, step = 20301 (0.177 sec)\nI0809 20:42:46.779720 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 553.116\nI0809 20:42:46.780584 139715572925888 basic_session_run_hooks.py:260] loss = 32.721966, step = 20401 (0.181 sec)\nI0809 20:42:46.975269 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 511.355\nI0809 20:42:46.976063 139715572925888 basic_session_run_hooks.py:260] loss = 37.798504, step = 20501 (0.195 sec)\nI0809 20:42:47.161967 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 535.676\nI0809 20:42:47.162949 139715572925888 basic_session_run_hooks.py:260] loss = 78.754654, step = 20601 (0.187 sec)\nI0809 20:42:47.364392 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 493.978\nI0809 20:42:47.365150 139715572925888 basic_session_run_hooks.py:260] loss = 25.544836, step = 20701 (0.202 sec)\nI0809 20:42:47.564527 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 499.706\nI0809 20:42:47.565649 139715572925888 basic_session_run_hooks.py:260] loss = 75.64566, step = 20801 (0.200 sec)\nI0809 20:42:47.746356 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 549.946\nI0809 20:42:47.747339 139715572925888 basic_session_run_hooks.py:260] loss = 26.593527, step = 20901 (0.182 sec)\nI0809 20:42:47.927987 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 550.525\nI0809 20:42:47.928855 139715572925888 basic_session_run_hooks.py:260] loss = 61.141468, step = 21001 (0.182 sec)\nI0809 20:42:48.110887 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 546.81\nI0809 20:42:48.111720 139715572925888 basic_session_run_hooks.py:260] loss = 30.508247, step = 21101 (0.183 sec)\nI0809 20:42:48.291301 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 554.299\nI0809 20:42:48.292495 139715572925888 basic_session_run_hooks.py:260] loss = 40.44137, step = 21201 (0.181 sec)\nI0809 20:42:48.477852 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 535.969\nI0809 20:42:48.478656 139715572925888 basic_session_run_hooks.py:260] loss = 37.764538, step = 21301 (0.186 sec)\nI0809 20:42:48.680670 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 493.08\nI0809 20:42:48.681519 139715572925888 basic_session_run_hooks.py:260] loss = 12.602806, step = 21401 (0.203 sec)\nI0809 20:42:48.887533 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 483.386\nI0809 20:42:48.888473 139715572925888 basic_session_run_hooks.py:260] loss = 66.86643, step = 21501 (0.207 sec)\nI0809 20:42:49.094826 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 482.493\nI0809 20:42:49.095681 139715572925888 basic_session_run_hooks.py:260] loss = 26.889442, step = 21601 (0.207 sec)\nI0809 20:42:49.303699 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 478.714\nI0809 20:42:49.304606 139715572925888 basic_session_run_hooks.py:260] loss = 34.189625, step = 21701 (0.209 sec)\nI0809 20:42:49.505270 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 496.374\nI0809 20:42:49.505960 139715572925888 basic_session_run_hooks.py:260] loss = 27.283232, step = 21801 (0.201 sec)\nI0809 20:42:49.687783 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 547.603\nI0809 20:42:49.688620 139715572925888 basic_session_run_hooks.py:260] loss = 25.419231, step = 21901 (0.183 sec)\nI0809 20:42:49.893177 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 486.855\nI0809 20:42:49.893961 139715572925888 basic_session_run_hooks.py:260] loss = 51.8715, step = 22001 (0.205 sec)\nI0809 20:42:50.094173 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 497.505\nI0809 20:42:50.094955 139715572925888 basic_session_run_hooks.py:260] loss = 55.552635, step = 22101 (0.201 sec)\nI0809 20:42:50.284781 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 524.643\nI0809 20:42:50.285800 139715572925888 basic_session_run_hooks.py:260] loss = 21.97137, step = 22201 (0.191 sec)\nI0809 20:42:50.479226 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 514.31\nI0809 20:42:50.480102 139715572925888 basic_session_run_hooks.py:260] loss = 18.18085, step = 22301 (0.194 sec)\nI0809 20:42:50.657841 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 560.096\nI0809 20:42:50.658891 139715572925888 basic_session_run_hooks.py:260] loss = 56.170902, step = 22401 (0.179 sec)\nI0809 20:42:50.835670 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 562.079\nI0809 20:42:50.836515 139715572925888 basic_session_run_hooks.py:260] loss = 44.311165, step = 22501 (0.178 sec)\nI0809 20:42:51.014305 139715572925888 basic_session_run_hooks.py:692] global_step/sec: 559.767\nI0809 20:42:51.015145 139715572925888 basic_session_run_hooks.py:260] loss = 46.684082, step = 22601 (0.179 sec)\nI0809 20:42:51.116461 139715572925888 basic_session_run_hooks.py:606] Saving checkpoints for 22650 into house_trained/model.ckpt.\nI0809 20:42:51.181637 139715572925888 estimator.py:1145] Calling model_fn.\nI0809 20:42:51.181864 139715572925888 estimator.py:1145] Calling model_fn.\nI0809 20:42:51.391630 139715572925888 estimator.py:1147] Done calling model_fn.\nW0809 20:42:51.392601 139715572925888 deprecation_wrapper.py:119] From /home/jupyter/training-data-analyst/courses/machine_learning/deepdive/05_artandscience/house_prediction_module/trainer/model.py:50: The name tf.metrics.root_mean_squared_error is deprecated. Please use tf.compat.v1.metrics.root_mean_squared_error instead.\n\nI0809 20:42:51.410948 139715572925888 estimator.py:1147] Done calling model_fn.\nI0809 20:42:51.430068 139715572925888 evaluation.py:255] Starting evaluation at 2019-08-09T20:42:51Z\nI0809 20:42:51.510735 139715572925888 monitored_session.py:240] Graph was finalized.\nW0809 20:42:51.511337 139715572925888 deprecation.py:323] From /usr/local/lib/python2.7/dist-packages/tensorflow/python/training/saver.py:1276: checkpoint_exists (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse standard file APIs to check for files with this prefix.\nI0809 20:42:51.512527 139715572925888 saver.py:1280] Restoring parameters from house_trained/model.ckpt-22650\nI0809 20:42:51.556935 139715572925888 session_manager.py:500] Running local_init_op.\nI0809 20:42:51.588593 139715572925888 session_manager.py:502] Done running local_init_op.\nI0809 20:42:51.886137 139715572925888 evaluation.py:275] Finished evaluation at 2019-08-09-20:42:51\nI0809 20:42:51.886435 139715572925888 estimator.py:2039] Saving dict for global step 22650: average_loss = 1.2751312, global_step = 22650, label/mean = 2.0454624, loss = 4320.1445, prediction/mean = 2.022154, rmse = 112921.7\nI0809 20:42:51.945333 139715572925888 estimator.py:2099] Saving 'checkpoint_path' summary for global step 22650: house_trained/model.ckpt-22650\nI0809 20:42:51.982523 139715572925888 estimator.py:368] Loss for final step: 33.386158.\n"
]
],
[
[
"# Create hyperparam.yaml",
"_____no_output_____"
]
],
[
[
"%%writefile hyperparam.yaml\ntrainingInput:\n hyperparameters:\n goal: MINIMIZE\n maxTrials: 5\n maxParallelTrials: 1\n hyperparameterMetricTag: rmse\n params:\n - parameterName: batch_size\n type: INTEGER\n minValue: 8\n maxValue: 64\n scaleType: UNIT_LINEAR_SCALE\n - parameterName: learning_rate\n type: DOUBLE\n minValue: 0.01\n maxValue: 0.1\n scaleType: UNIT_LOG_SCALE",
"Writing hyperparam.yaml\n"
],
[
"%%bash\nOUTDIR=gs://${BUCKET}/house_trained # CHANGE bucket name appropriately\ngsutil rm -rf $OUTDIR\nexport PYTHONPATH=${PYTHONPATH}:${PWD}/house_prediction_module\ngcloud ai-platform jobs submit training house_$(date -u +%y%m%d_%H%M%S) \\\n --config=hyperparam.yaml \\\n --module-name=trainer.task \\\n --package-path=$(pwd)/house_prediction_module/trainer \\\n --job-dir=$OUTDIR \\\n --runtime-version=$TFVERSION \\\n --\\\n --output_dir=$OUTDIR \\",
"jobId: house_190809_204253\nstate: QUEUED\n"
],
[
"!gcloud ai-platform jobs describe house_190809_204253 # CHANGE jobId appropriately",
"createTime: '2019-08-09T20:42:55Z'\netag: zU1W9lhyf0w=\njobId: house_190809_204253\nstartTime: '2019-08-09T20:42:59Z'\nstate: RUNNING\ntrainingInput:\n args:\n - --output_dir=gs://qwiklabs-gcp-faf328caac1ef9a0/house_trained\n hyperparameters:\n goal: MINIMIZE\n hyperparameterMetricTag: rmse\n maxParallelTrials: 1\n maxTrials: 5\n params:\n - maxValue: 64.0\n minValue: 8.0\n parameterName: batch_size\n scaleType: UNIT_LINEAR_SCALE\n type: INTEGER\n - maxValue: 0.1\n minValue: 0.01\n parameterName: learning_rate\n scaleType: UNIT_LOG_SCALE\n type: DOUBLE\n jobDir: gs://qwiklabs-gcp-faf328caac1ef9a0/house_trained\n packageUris:\n - gs://qwiklabs-gcp-faf328caac1ef9a0/house_trained/packages/2148c5e4ea8c7f8c90ee6fdaffa93a2f5fce6ef0bdb95b679c1067e97d0f01e7/trainer-0.0.0.tar.gz\n pythonModule: trainer.task\n region: us-east1\n runtimeVersion: '1.8'\ntrainingOutput:\n hyperparameterMetricTag: rmse\n isHyperparameterTuningJob: true\n\nView job in the Cloud Console at:\nhttps://console.cloud.google.com/mlengine/jobs/house_190809_204253?project=qwiklabs-gcp-faf328caac1ef9a0\n\nView logs at:\nhttps://console.cloud.google.com/logs?resource=ml.googleapis.com%2Fjob_id%2Fhouse_190809_204253&project=qwiklabs-gcp-faf328caac1ef9a0\n\n\nTo take a quick anonymous survey, run:\n $ gcloud alpha survey\n\n"
]
],
[
[
"## Challenge exercise\nAdd a few engineered features to the housing model, and use hyperparameter tuning to choose which set of features the model uses.\n\n<p>\nCopyright 2018 Google Inc. Licensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
d00373e5b51e057071a5b04a77473d2273319501 | 47,013 | ipynb | Jupyter Notebook | semrep/evaluate/koehn/koehn.ipynb | geoffbacon/semrep | 366d5740a117f47cda73807a8b9e6b7cf1ca8138 | [
"MIT"
] | null | null | null | semrep/evaluate/koehn/koehn.ipynb | geoffbacon/semrep | 366d5740a117f47cda73807a8b9e6b7cf1ca8138 | [
"MIT"
] | null | null | null | semrep/evaluate/koehn/koehn.ipynb | geoffbacon/semrep | 366d5740a117f47cda73807a8b9e6b7cf1ca8138 | [
"MIT"
] | null | null | null | 51.890728 | 10,328 | 0.584349 | [
[
[
"# Köhn\n\nIn this notebook I replicate Koehn (2015): _What's in an embedding? Analyzing word embeddings through multilingual evaluation_. This paper proposes to i) evaluate an embedding method on more than one language, and ii) evaluate an embedding model by how well its embeddings capture syntactic features. He uses an L2-regularized linear classifier, with an upper baseline that assigns the most frequent class. He finds that most methods perform similarly on this task, but that dependency based embeddings perform better. Dependency based embeddings particularly perform better when you decrease the dimensionality. Overall, the aim is to have an evalation method that tells you something about the structure of the learnt representations. He evaulates a range of different models on their ability to capture a number of different morphosyntactic features in a bunch of languages.\n\n**Embedding models tested:**\n- cbow\n- skip-gram\n- glove\n- dep\n- cca\n- brown\n\n**Features tested:**\n- pos\n- headpos (the pos of the word's head)\n- label\n- gender\n- case\n- number\n- tense\n\n**Languages tested:**\n- Basque\n- English\n- French\n- German\n- Hungarian\n- Polish\n- Swedish\n\nWord embeddings were trained on automatically PoS-tagged and dependency-parsed data using existing models. This is so the dependency-based embeddings can be trained. The evaluation is on hand-labelled data. English training data is a subset of Wikipedia; English test data comes from PTB. For all other languages, both the training and test data come from a shared task on parsing morphologically rich languages. Koehn trained embeddings with window size 5 and 11 and dimensionality 10, 100, 200.\n\nDependency-based embeddings perform the best on almost all tasks. They even do well when the dimensionality is reduced to 10, while other methods perform poorly in this case.\n\nI'll need:\n- models\n- learnt representations\n- automatically labeled data\n- hand-labeled data",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport os\nimport csv\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()\nfrom sklearn.linear_model import LogisticRegression, LogisticRegressionCV\nfrom sklearn.model_selection import train_test_split, StratifiedKFold\nfrom sklearn.metrics import roc_curve, roc_auc_score, classification_report, confusion_matrix\nfrom sklearn.preprocessing import LabelEncoder\n\ndata_path = '../../data'\ntmp_path = '../../tmp'",
"_____no_output_____"
]
],
[
[
"## Learnt representations\n\n### GloVe",
"_____no_output_____"
]
],
[
[
"size = 50\nfname = 'embeddings/glove.6B.{}d.txt'.format(size)\nglove_path = os.path.join(data_path, fname)\nglove = pd.read_csv(glove_path, sep=' ', header=None, index_col=0, quoting=csv.QUOTE_NONE)\nglove.head()",
"_____no_output_____"
]
],
[
[
"## Features",
"_____no_output_____"
]
],
[
[
"fname = 'UD_English/features.csv'\nfeatures_path = os.path.join(data_path, os.path.join('evaluation/dependency', fname))\nfeatures = pd.read_csv(features_path).set_index('form')\nfeatures.head()",
"/home/bacon/miniconda3/lib/python3.6/site-packages/IPython/core/interactiveshell.py:2698: DtypeWarning: Columns (7) have mixed types. Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
],
[
"df = pd.merge(glove, features, how='inner', left_index=True, right_index=True)\ndf.head()",
"_____no_output_____"
]
],
[
[
"## Prediction",
"_____no_output_____"
]
],
[
[
"def prepare_X_and_y(feature, data):\n \"\"\"Return X and y ready for predicting feature from embeddings.\"\"\"\n relevant_data = data[data[feature].notnull()]\n columns = list(range(1, size+1))\n X = relevant_data[columns]\n y = relevant_data[feature]\n train = relevant_data['set'] == 'train'\n test = (relevant_data['set'] == 'test') | (relevant_data['set'] == 'dev')\n X_train, X_test = X[train].values, X[test].values\n y_train, y_test = y[train].values, y[test].values\n return X_train, X_test, y_train, y_test\n\ndef predict(model, X_test):\n \"\"\"Wrapper for getting predictions.\"\"\"\n results = model.predict_proba(X_test)\n return np.array([t for f,t in results]).reshape(-1,1)\n\ndef conmat(model, X_test, y_test):\n \"\"\"Wrapper for sklearn's confusion matrix.\"\"\"\n y_pred = model.predict(X_test)\n c = confusion_matrix(y_test, y_pred)\n sns.heatmap(c, annot=True, fmt='d', \n xticklabels=model.classes_, \n yticklabels=model.classes_, \n cmap=\"YlGnBu\", cbar=False)\n plt.ylabel('Ground truth')\n plt.xlabel('Prediction')\n\ndef draw_roc(model, X_test, y_test):\n \"\"\"Convenience function to draw ROC curve.\"\"\"\n y_pred = predict(model, X_test)\n fpr, tpr, thresholds = roc_curve(y_test, y_pred)\n roc = roc_auc_score(y_test, y_pred)\n label = r'$AUC={}$'.format(str(round(roc, 3)))\n plt.plot(fpr, tpr, label=label);\n plt.title('ROC')\n plt.xlabel('False positive rate');\n plt.ylabel('True positive rate');\n plt.legend();\n\ndef cross_val_auc(model, X, y):\n for _ in range(5):\n X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, stratify=y)\n model = model.fit(X_train, y_train)\n draw_roc(model, X_test, y_test)",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = prepare_X_and_y('Tense', df)\n\nmodel = LogisticRegression(penalty='l2', solver='liblinear')\nmodel = model.fit(X_train, y_train)\nconmat(model, X_test, y_test)",
"_____no_output_____"
],
[
"sns.distplot(model.coef_[0], rug=True, kde=False);",
"_____no_output_____"
]
],
[
[
"# Hyperparameter optimization before error analysis",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
d00374ac930c1e739d6267bffe828dc3276c1021 | 52,174 | ipynb | Jupyter Notebook | Course 2 - CNNs in Tensorflow/Exercise5_CatsDogs.ipynb | chrismartinis/tensorflow_developer_professional_certificate | e81bdc75b3b3cbd46911ca816445fa07061dcb7a | [
"MIT"
] | 5 | 2020-09-16T15:53:00.000Z | 2021-05-23T08:53:59.000Z | Course 2 - CNNs in Tensorflow/Exercise5_CatsDogs.ipynb | Ambiyang/tensorflow_developer_professional_certificate | e81bdc75b3b3cbd46911ca816445fa07061dcb7a | [
"MIT"
] | null | null | null | Course 2 - CNNs in Tensorflow/Exercise5_CatsDogs.ipynb | Ambiyang/tensorflow_developer_professional_certificate | e81bdc75b3b3cbd46911ca816445fa07061dcb7a | [
"MIT"
] | 7 | 2020-10-18T12:06:50.000Z | 2021-10-06T13:20:43.000Z | 85.953871 | 14,872 | 0.7493 | [
[
[
"# In this exercise you will train a CNN on the FULL Cats-v-dogs dataset\n# This will require you doing a lot of data preprocessing because\n# the dataset isn't split into training and validation for you\n# This code block has all the required inputs\nimport os\nimport zipfile\nimport random\nimport tensorflow as tf\nfrom tensorflow.keras.optimizers import RMSprop\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\nfrom shutil import copyfile",
"_____no_output_____"
],
[
"# This code block downloads the full Cats-v-Dogs dataset and stores it as \n# cats-and-dogs.zip. It then unzips it to /tmp\n# which will create a tmp/PetImages directory containing subdirectories\n# called 'Cat' and 'Dog' (that's how the original researchers structured it)\n# If the URL doesn't work, \n# . visit https://www.microsoft.com/en-us/download/confirmation.aspx?id=54765\n# And right click on the 'Download Manually' link to get a new URL\n\n!wget --no-check-certificate \\\n \"https://download.microsoft.com/download/3/E/1/3E1C3F21-ECDB-4869-8368-6DEBA77B919F/kagglecatsanddogs_3367a.zip\" \\\n -O \"/tmp/cats-and-dogs.zip\"\n\nlocal_zip = '/tmp/cats-and-dogs.zip'\nzip_ref = zipfile.ZipFile(local_zip, 'r')\nzip_ref.extractall('/tmp')\nzip_ref.close()\n",
"--2020-02-06 09:38:28-- https://download.microsoft.com/download/3/E/1/3E1C3F21-ECDB-4869-8368-6DEBA77B919F/kagglecatsanddogs_3367a.zip\nResolving download.microsoft.com (download.microsoft.com)... 2.21.40.213, 2a02:26f0:6b:5b3::e59, 2a02:26f0:6b:5a5::e59\nConnecting to download.microsoft.com (download.microsoft.com)|2.21.40.213|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 824894548 (787M) [application/octet-stream]\nSaving to: ‘/tmp/cats-and-dogs.zip’\n\n/tmp/cats-and-dogs. 100%[===================>] 786.68M 294MB/s in 2.7s \n\n2020-02-06 09:38:31 (294 MB/s) - ‘/tmp/cats-and-dogs.zip’ saved [824894548/824894548]\n\n"
],
[
"print(len(os.listdir('/tmp/PetImages/Cat/')))\nprint(len(os.listdir('/tmp/PetImages/Dog/')))\n\n# Expected Output:\n# 12501\n# 12501",
"12501\n12501\n"
],
[
"# Use os.mkdir to create your directories\n# You will need a directory for cats-v-dogs, and subdirectories for training\n# and testing. These in turn will need subdirectories for 'cats' and 'dogs'\ntry:\n os.mkdir(\"/tmp/cats-v-dogs\")\n os.mkdir(\"/tmp/cats-v-dogs/training\")\n os.mkdir(\"/tmp/cats-v-dogs/testing\")\n os.mkdir(\"/tmp/cats-v-dogs/training/dogs\")\n os.mkdir(\"/tmp/cats-v-dogs/training/cats\")\n os.mkdir(\"/tmp/cats-v-dogs/testing/dogs\")\n os.mkdir(\"/tmp/cats-v-dogs/testing/cats\")\nexcept OSError:\n pass",
"_____no_output_____"
],
[
"# Write a python function called split_data which takes\n# a SOURCE directory containing the files\n# a TRAINING directory that a portion of the files will be copied to\n# a TESTING directory that a portion of the files will be copied to\n# a SPLIT SIZE to determine the portion\n# The files should also be randomized, so that the training set is a random\n# X% of the files, and the test set is the remaining files\n# SO, for example, if SOURCE is PetImages/Cat, and SPLIT SIZE is .9\n# Then 90% of the images in PetImages/Cat will be copied to the TRAINING dir\n# and 10% of the images will be copied to the TESTING dir\n# Also -- All images should be checked, and if they have a zero file length,\n# they will not be copied over\n#\n# os.listdir(DIRECTORY) gives you a listing of the contents of that directory\n# os.path.getsize(PATH) gives you the size of the file\n# copyfile(source, destination) copies a file from source to destination\n# random.sample(list, len(list)) shuffles a list\ndef split_data(SOURCE, TRAINING, TESTING, SPLIT_SIZE):\n files = []\n for filename in os.listdir(SOURCE):\n file = SOURCE + filename\n if os.path.getsize(file) > 0:\n files.append(filename)\n else:\n print (filename + \" is zero length, so ignoring.\")\n\n training_length = int(len(files) * SPLIT_SIZE)\n testing_length = int(len(files) - training_length)\n shuffled_set = random.sample(files, len(files))\n training_set = shuffled_set[0:training_length]\n testing_set = shuffled_set[-testing_length:]\n\n for filename in training_set:\n src = SOURCE + filename\n dst = TRAINING + filename\n copyfile(src, dst)\n\n for filename in testing_set:\n src = SOURCE + filename\n dst = TESTING + filename\n copyfile(src, dst)\n\n\nCAT_SOURCE_DIR = \"/tmp/PetImages/Cat/\"\nTRAINING_CATS_DIR = \"/tmp/cats-v-dogs/training/cats/\"\nTESTING_CATS_DIR = \"/tmp/cats-v-dogs/testing/cats/\"\nDOG_SOURCE_DIR = \"/tmp/PetImages/Dog/\"\nTRAINING_DOGS_DIR = \"/tmp/cats-v-dogs/training/dogs/\"\nTESTING_DOGS_DIR = \"/tmp/cats-v-dogs/testing/dogs/\"\n\nsplit_size = .9\nsplit_data(CAT_SOURCE_DIR, TRAINING_CATS_DIR, TESTING_CATS_DIR, split_size)\nsplit_data(DOG_SOURCE_DIR, TRAINING_DOGS_DIR, TESTING_DOGS_DIR, split_size)\n\n# Expected output\n# 666.jpg is zero length, so ignoring\n# 11702.jpg is zero length, so ignoring",
"666.jpg is zero length, so ignoring.\n11702.jpg is zero length, so ignoring.\n"
],
[
"print(len(os.listdir('/tmp/cats-v-dogs/training/cats/')))\nprint(len(os.listdir('/tmp/cats-v-dogs/training/dogs/')))\nprint(len(os.listdir('/tmp/cats-v-dogs/testing/cats/')))\nprint(len(os.listdir('/tmp/cats-v-dogs/testing/dogs/')))\n\n# Expected output:\n# 11250\n# 11250\n# 1250\n# 1250",
"12371\n12352\n2371\n2352\n"
],
[
"# DEFINE A KERAS MODEL TO CLASSIFY CATS V DOGS\n# USE AT LEAST 3 CONVOLUTION LAYERS\nmodel = tf.keras.models.Sequential([\n tf.keras.layers.Conv2D(16, (3,3), activation='relu', input_shape=(150,150,3)),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Conv2D(32, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Conv2D(64, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(512, activation='relu'),\n tf.keras.layers.Dense(1, activation='sigmoid')\n])\n\nmodel.compile(optimizer=RMSprop(lr=0.001),\n loss='binary_crossentropy',\n metrics=['acc'])",
"_____no_output_____"
],
[
"TRAINING_DIR = \"/tmp/cats-v-dogs/training/\"\ntrain_datagen = ImageDataGenerator(rescale=1./255)\ntrain_generator = train_datagen.flow_from_directory(TRAINING_DIR,\n batch_size=100,\n class_mode='binary',\n target_size=(150,150))\n\nVALIDATION_DIR = \"/tmp/cats-v-dogs/testing/\"\nvalidation_datagen = ImageDataGenerator(rescale=1./255)\nvalidation_generator = train_datagen.flow_from_directory(VALIDATION_DIR,\n batch_size=10,\n class_mode='binary',\n target_size=(150,150))\n\n\n\n# Expected Output:\n# Found 22498 images belonging to 2 classes.\n# Found 2500 images belonging to 2 classes.",
"Found 24721 images belonging to 2 classes.\nFound 4723 images belonging to 2 classes.\n"
],
[
"history = model.fit_generator(train_generator,\n epochs=15,\n verbose=1,\n validation_data=validation_generator)\n\n# The expectation here is that the model will train, and that accuracy will be > 95% on both training and validation\n# i.e. acc:A1 and val_acc:A2 will be visible, and both A1 and A2 will be > .9",
"Epoch 1/15\n 54/248 [=====>........................] - ETA: 53s - loss: 0.8276 - acc: 0.5615"
],
[
"# PLOT LOSS AND ACCURACY\n%matplotlib inline\n\nimport matplotlib.image as mpimg\nimport matplotlib.pyplot as plt\n\n#-----------------------------------------------------------\n# Retrieve a list of list results on training and test data\n# sets for each training epoch\n#-----------------------------------------------------------\nacc=history.history['acc']\nval_acc=history.history['val_acc']\nloss=history.history['loss']\nval_loss=history.history['val_loss']\n\nepochs=range(len(acc)) # Get number of epochs\n\n#------------------------------------------------\n# Plot training and validation accuracy per epoch\n#------------------------------------------------\nplt.plot(epochs, acc, 'r', \"Training Accuracy\")\nplt.plot(epochs, val_acc, 'b', \"Validation Accuracy\")\nplt.title('Training and validation accuracy')\nplt.figure()\n\n#------------------------------------------------\n# Plot training and validation loss per epoch\n#------------------------------------------------\nplt.plot(epochs, loss, 'r', \"Training Loss\")\nplt.plot(epochs, val_loss, 'b', \"Validation Loss\")\n\n\nplt.title('Training and validation loss')\n\n# Desired output. Charts with training and validation metrics. No crash :)",
"_____no_output_____"
],
[
"# Here's a codeblock just for fun. You should be able to upload an image here \n# and have it classified without crashing\n\nimport numpy as np\nfrom google.colab import files\nfrom keras.preprocessing import image\n\nuploaded = files.upload()\n\nfor fn in uploaded.keys():\n \n # predicting images\n path = '/content/' + fn\n img = image.load_img(path, target_size=(# YOUR CODE HERE))\n x = image.img_to_array(img)\n x = np.expand_dims(x, axis=0)\n\n images = np.vstack([x])\n classes = model.predict(images, batch_size=10)\n print(classes[0])\n if classes[0]>0.5:\n print(fn + \" is a dog\")\n else:\n print(fn + \" is a cat\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0037ca8fffe500d5e12ef7e2fb229299b5b3079 | 325,870 | ipynb | Jupyter Notebook | Multi-armed Bandits.ipynb | yash-deshpande/decorrelating-linear-models | a638f07ceba3ec700ef53aba5579fe9fefa7ea6a | [
"MIT"
] | 4 | 2018-12-25T18:51:58.000Z | 2021-06-17T05:33:33.000Z | Multi-armed Bandits.ipynb | yash-deshpande/decorrelating-linear-models | a638f07ceba3ec700ef53aba5579fe9fefa7ea6a | [
"MIT"
] | null | null | null | Multi-armed Bandits.ipynb | yash-deshpande/decorrelating-linear-models | a638f07ceba3ec700ef53aba5579fe9fefa7ea6a | [
"MIT"
] | 2 | 2019-06-17T06:52:51.000Z | 2020-06-24T13:00:16.000Z | 291.215371 | 31,888 | 0.892785 | [
[
[
"# Confidence interval and bias comparison in the multi-armed bandit\n# setting of https://arxiv.org/pdf/1507.08025.pdf\nimport numpy as np\nimport pandas as pd\nimport scipy.stats as stats\nimport time\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport seaborn as sns\nsns.set(style='white', palette='colorblind', color_codes=True)\n",
"_____no_output_____"
],
[
"#\n# Experiment parameters\n#\n# Set random seed for reproducibility\nseed = 1234\nnp.random.seed(seed)\n# Trial repetitions (number of times experiment is repeated)\nR = 5000\n# Trial size (total number of arm pulls)\nT = 1000\n# Number of arms\nK = 2\n# Noise distribution: 2*beta(alph, alph) - 1\nnoise_param = 1.0 # uniform distribution\n# Parameters of Gaussian distribution prior on each arm\nmu0 = 0.4 # prior mean\nvar0 = 1/(2*noise_param + 1.0) # prior variance set to correct value\n# Select reward means for each arm and set variance\nreward_means = np.concatenate([np.repeat(.3, K-1), [.30]])\nreward_vars = np.repeat(var0, K)\n# Select probability of choosing current belief in epsilon greedy policy\nECB_epsilon = .1\n",
"_____no_output_____"
],
[
"#\n# Evaluation parameters\n#\n# Confidence levels for confidence regions\nconfidence_levels = np.arange(0.9, 1.0, step=0.01)\n# Standard normal error thresholds for two-sided (univariate) intervals with given confidence level\ngaussian_thresholds_ts = -stats.norm.ppf((1.0-confidence_levels)/2.0)\ngaussian_thresholds_os = -stats.norm.ppf(1.0-confidence_levels)\nprint gaussian_thresholds_ts\nprint gaussian_thresholds_os",
"[ 1.64485363 1.69539771 1.75068607 1.81191067 1.88079361 1.95996398\n 2.05374891 2.17009038 2.32634787 2.5758293 ]\n[ 1.28155157 1.34075503 1.40507156 1.47579103 1.55477359 1.64485363\n 1.75068607 1.88079361 2.05374891 2.32634787]\n"
],
[
"#\n# Define arm selection policies\n#\npolicies = {}\n# Epsilon-greedy: select current belief (arm with highest posterior reward \n# probability) w.p. 1-epsilon and arm uniformly at random otherwise\ndef ECB(mu_post, var_post, epsilon=ECB_epsilon):\n # Determine whether to select current belief by flipping biased coin\n use_cb = np.random.binomial(1, 1.0-epsilon)\n if use_cb:\n # Select arm with highest posterior reward probability\n arm = np.argmax(mu_post)\n else:\n # Select arm uniformly at random\n arm = np.random.choice(xrange(K))\n return arm\npolicies['ECB'] = ECB\n# Current belief: select arm with highest posterior probability\ndef CB(mu_post, var_post):\n return ECB(mu_post, var_post, epsilon=0.0)\n# policies['CB'] = CB\n# Fixed randomized design: each arm selected independently and uniformly\ndef FR(mu_post, var_post, epsilon=ECB_epsilon):\n return ECB(mu_post, var_post, epsilon=1.0)\npolicies['FR'] = FR\n# Thompson sampling: select arm k with probability proportional to P(arm k has highest reward | data)^c\n# where c = 1 and P(arm k has highest reward | data) is the posterior probability that arm k has\n# the highest reward\n# TODO: the paper uses c = t/(2T) instead, citing Thall and Wathen (2007); investigate how to achieve this efficiently\ndef TS(mu_post, var_post, epsilon=ECB_epsilon):\n # Draw a sample from each arm's posterior\n samples = np.random.normal(mu_post, np.sqrt(var_post))\n # Select an arm with the largest sample\n arm = np.argmax(samples)\n return arm\npolicies['TS'] = TS\ndef lilUCB(mu_post, var_post, epsilon=ECB_epsilon ):\n #define lilUCB params, see Jamieson et al 2013\n # use 1/variance as number of times the arm is tried.\n # at time t, choose arm k that maximizes:\n # muhat_k(t) + (1+beta)*(1+sqrt(eps))*sqrt{2(1+eps)/T_k}*sqrt{log(1/delta) + log(log((1+eps)*T_k))}\n # where muhat_k (t) is sample mean of k^th arm at time t and T_k = T_k(t) is the number of times arm k is tried\n # up toa time t\n epsilonUCB = 0.01\n betaUCB = 0.5\n aUCB = 1+ 2/betaUCB\n deltaUCB = 0.01\n \n lilFactorUCB = np.log(1/deltaUCB) + np.log(np.log((1+epsilonUCB)/var_post))\n scoresUCB = mu_post + (1+betaUCB)*(1+np.sqrt(epsilonUCB))*np.sqrt((2+2*epsilonUCB)*lilFactorUCB*var_post)\n arm = np.argmax(scoresUCB)\n return arm\npolicies['UCB'] = lilUCB",
"_____no_output_____"
],
[
"#\n# Gather data: Generate arm pulls and rewards using different policies\n#\ntic = time.time()\narms = []\nrewards = []\nfor r in xrange(R):\n arms.append(pd.DataFrame(index=range(0,T)))\n rewards.append(pd.DataFrame(index=range(0,T)))\n \n # Keep track of posterior beta parameters for each arm\n mu_post = np.repeat(mu0, K)\n var_post = np.repeat(var0, K)\n for policy in policies.keys():\n # Ensure arms column has integer type by initializing with integer value\n arms[r][policy] = 0\n for t in range(T):\n if t < K:\n # Ensure each arm selected at least once\n arm = t\n else:\n # Select an arm according to policy\n arm = policies[policy](mu_post, var_post, epsilon = ECB_epsilon)\n # Collect reward from selected arm\n reward = 2*np.random.beta(noise_param, noise_param) - 1.0 + reward_means[arm]\n # Update Gaussian posterior\n new_var = 1.0/(1.0/var_post[arm] + 1.0/reward_vars[arm])\n mu_post[arm] = (mu_post[arm]/var_post[arm] + reward/reward_vars[arm])*new_var\n var_post[arm] = new_var\n # Store results\n arms[r].set_value(t, policy, arm)\n rewards[r].set_value(t, policy, reward)\n \nprint \"{}s elapsed\".format(time.time()-tic)",
"401.451577187s elapsed\n"
],
[
"\n# Inspect arm selections\nprint arms[0][0:min(10,T)]",
" UCB FR ECB TS\n0 0 0 0 0\n1 1 1 1 1\n2 0 0 0 1\n3 1 0 1 1\n4 0 0 1 1\n5 0 1 1 1\n6 1 1 1 1\n7 1 1 1 1\n8 1 1 1 1\n9 1 0 1 0\n"
],
[
"# Display some summary statistics for the collected data\npct_arm_counts={}\nfor policy in arms[0].keys():\n print policy\n pct_arm_counts[policy] = np.percentile([arms[r][policy].groupby(arms[r][policy]).size().values \\\n for r in xrange(R)],15, axis=0)\npct_arm_counts ",
"UCB\nFR\nECB\nTS\n"
],
[
"# compute statistics for arm distributions\nn_arm1 = {}\nfor policy in policies:\n n_arm1[policy] = np.zeros(R)\nfor ix, run in enumerate(arms):\n for policy in policies:\n n_arm1[policy][ix] = sum(run[policy])",
"_____no_output_____"
],
[
"#plot histograms of arm distributions for each policy\n\npolicies = ['UCB', 'ECB', 'TS']\n\npolicy = 'ECB'\nfor ix, policy in enumerate(policies):\n fig, ax = plt.subplots(1, figsize=(5.5, 4))\n ax.set_title(policy, fontsize=title_font_size, fontweight='bold')\n sns.distplot(n_arm1[policy]/T, \n kde=False,\n bins=20,\n norm_hist=True,\n ax=ax, \n hist_kws=dict(alpha=0.8)\n )\n\n fig.savefig(path+'mab_{}_armdist'.format(policy))\n plt.show()\n",
"_____no_output_____"
],
[
"# \n# Form estimates: For each method, compute reward probability estimates and \n# single-parameter error thresholds for confidence intervals\n#\ntic = time.time()\nestimates = []\nthresholds_ts = []\nthresholds_os = []\nnormalized_errors = []\nfor r in xrange(R):\n estimates.append({})\n thresholds_ts.append({})\n thresholds_os.append({})\n normalized_errors.append({})\n for policy in arms[r].columns:\n # Create list of estimates and confidence regions for this policy\n estimates[r][policy] = {}\n thresholds_ts[r][policy] = {}\n thresholds_os[r][policy] = {}\n normalized_errors[r][policy] = {}\n \n # OLS with asymptotic Gaussian confidence\n #\n # Compute estimates of arm reward probabilities\n estimates[r][policy]['OLS_gsn'] = rewards[r][policy].groupby(arms[r][policy]).mean().values\n # Asymptotic marginal variances diag((X^tX)^{-1})\n arm_counts = arms[r][policy].groupby(arms[r][policy]).size().values\n variances = reward_vars / arm_counts\n # compute normalized errors\n normalized_errors[r][policy]['OLS_gsn'] = (estimates[r][policy]['OLS_gsn'] - reward_means)/np.sqrt(variances)\n \n # Compute asymptotic Gaussian single-parameter confidence thresholds\n thresholds_ts[r][policy]['OLS_gsn'] = np.outer(np.sqrt(variances), gaussian_thresholds_ts)\n thresholds_os[r][policy]['OLS_gsn'] = np.outer(np.sqrt(variances), gaussian_thresholds_os)\n #\n # OLS with concentration inequality confidence\n # \n # Compute estimates of arm reward probabilities\n estimates[r][policy]['OLS_conc'] = np.copy(estimates[r][policy]['OLS_gsn'])\n normalized_errors[r][policy]['OLS_conc'] = (estimates[r][policy]['OLS_gsn'] - reward_means)/np.sqrt(variances)\n\n # Compute single-parameter confidence intervals using concentration inequalities\n # of https://arxiv.org/pdf/1102.2670.pdf Sec. 4\n # threshold_ts = sqrt(reward_vars) * sqrt((1+N_k)/N_k^2 * (1+2*log(sqrt(1+N_k)/delta)))\n thresholds_ts[r][policy]['OLS_conc'] = np.sqrt(reward_vars/reward_vars)[:,None] * np.concatenate([ \n np.sqrt(((1.0+arm_counts)/arm_counts**2) * (1+2*np.log(np.sqrt(1.0+arm_counts)/(1-c))))[:,None]\n for c in confidence_levels], axis=1)\n thresholds_os[r][policy]['OLS_conc'] = np.copy(thresholds_ts[r][policy]['OLS_conc'])\n # \n # W estimate with asymptotic Gaussian confidence\n # Y: using lambda_min = min_median_arm_count/log(T) as W_Lambdas\n # avg_arm_counts = pct_arm_counts[policy]/log(T)\n W_lambdas = np.ones(T)*min(pct_arm_counts[policy])/np.log(T)\n # Latest parameter estimate vector\n beta = np.copy(estimates[r][policy]['OLS_gsn']) ###\n # Latest w_t vector\n w = np.zeros((K))\n # Latest matrix W_tX_t = w_1 x_1^T + ... + w_t x_t^T\n WX = np.zeros((K,K))\n # Latest vector of marginal variances reward_vars * (w_1**2 + ... + w_t**2)\n variances = np.zeros(K)\n for t in range(T):\n # x_t = e_{arm}\n arm = arms[r][policy][t]\n # y_t = reward\n reward = rewards[r][policy][t]\n # Update w_t = (1/(norm{x_t}^2+lambda_t)) (x_t - W_{t-1} X_{t-1} x_t)\n np.copyto(w, -WX[:,arm])\n w[arm] += 1\n w /= (1.0+W_lambdas[t])\n # Update beta_t = beta_{t-1} + w_t (y_t - <beta_OLS, x_t>)\n beta += w * (reward - estimates[r][policy]['OLS_gsn'][arm]) ###\n # Update W_tX_t = W_{t-1}X_{t-1} + w_t x_t^T \n WX[:,arm] += w\n # Update marginal variances\n variances += reward_vars * w**2\n estimates[r][policy]['W'] = beta\n normalized_errors[r][policy]['W'] = (estimates[r][policy]['W'] - reward_means)/np.sqrt(variances)\n\n # Compute asymptotic Gaussian single-parameter confidence thresholds and coverage\n thresholds_ts[r][policy]['W'] = np.outer(np.sqrt(variances), gaussian_thresholds_ts)\n thresholds_os[r][policy]['W'] = np.outer(np.sqrt(variances), gaussian_thresholds_os)\nprint \"{}s elapsed\".format(time.time()-tic)\n",
"900.15949893s elapsed\n"
],
[
"# Display some summary statistics concerning the model estimates\nif False:\n for policy in [\"ECB\",\"TS\"]:#arms[0].keys():\n for method in estimates[0][policy].keys():\n print \"{} {}\".format(policy, method)\n print \"average estimate: {}\".format(np.mean([estimates[r][policy][method] for r in xrange(R)], axis=0))\n print \"average threshold:\\n{}\".format(np.mean([thresholds_os[r][policy][method] for r in xrange(R)], axis=0))\n print \"\"",
"_____no_output_____"
],
[
"#\n# Evaluate estimates: For each policy and method, compute confidence interval \n# coverage probability and width\n#\ntic = time.time()\ncoverage = [] # Check if truth in [estimate +/- thresh]\nupper_coverage = [] # Check if truth >= estimate - thresh\nlower_coverage = [] # Check if truth <= estimate + thresh\nupper_sum_coverage = [] # Check if beta_2 - beta_1 >= estimate - thresh\nlower_sum_coverage = [] # Check if beta_2 - beta_1 <= estimate + thresh\nsum_norm = [] # compute (betahat_2 - beta_2 - betahat_1 + beta_1 ) / sqrt(variance_2 + variance_1)\n\n\nfor r in xrange(R):\n coverage.append({})\n upper_coverage.append({})\n lower_coverage.append({})\n upper_sum_coverage.append({})\n lower_sum_coverage.append({})\n sum_norm.append({})\n for policy in estimates[r].keys():\n # Interval coverage for each method\n coverage[r][policy] = {}\n upper_coverage[r][policy] = {}\n lower_coverage[r][policy] = {}\n upper_sum_coverage[r][policy] = {}\n lower_sum_coverage[r][policy] = {}\n sum_norm[r][policy] = {}\n for method in estimates[r][policy].keys():\n # Compute error of estimate\n error = estimates[r][policy][method] - reward_means\n # compute normalized sum\n # first compute arm variances\n stddevs = thresholds_os[r][policy][method].dot(gaussian_thresholds_os)/gaussian_thresholds_os.dot(gaussian_thresholds_os)\n variances = stddevs**2\n sum_norm[r][policy][method] = (error[0] + error[1])/np.sqrt(variances[0] + variances[1]) \n # Compute coverage of interval\n coverage[r][policy][method] = np.absolute(error)[:,None] <= thresholds_ts[r][policy][method]\n upper_coverage[r][policy][method] = error[:,None] <= thresholds_os[r][policy][method]\n lower_coverage[r][policy][method] = error[:,None] >= -thresholds_os[r][policy][method]\n upper_sum_coverage[r][policy][method] = error[1]+error[0] <= np.sqrt((thresholds_os[r][policy][method]**2).sum(axis=0))\n lower_sum_coverage[r][policy][method] = error[1]+error[0] >= -np.sqrt((thresholds_os[r][policy][method]**2).sum(axis=0))\nprint \"{}s elapsed\".format(time.time()-tic)",
"2.79937911034s elapsed\n"
],
[
"# set up some plotting configuration\n\npath = 'figs/'\npolicies = ['UCB', 'TS', 'ECB']\nmethods = [\"OLS_gsn\",\"OLS_conc\", \"W\"]\nmarkers = {}\nmarkers['OLS_gsn'] = 'v'\nmarkers['OLS_conc'] = '^'\nmarkers['W'] = 'o'\n\ncolors = {}\ncolors['OLS_gsn'] = sns.color_palette()[0]\ncolors['OLS_conc'] = sns.color_palette()[2]\ncolors['W'] = sns.color_palette()[1]\ncolors['Nominal'] = (0, 0, 0)\ncolors['OLS_emp'] = sns.color_palette()[3]\n\nlegend_font_size = 14\nlabel_font_size = 14\ntitle_font_size = 16\n",
"_____no_output_____"
],
[
"#\n# Display coverage results\n#\n## Select coverage array from {\"coverage\", \"lower_coverage\", \"upper_coverage\"}\n#coverage_type = \"lower_coverage\"\n#coverage_arr = locals()[coverage_type]\n# For each policy and method, display coverage as a function of confidence level\n\nmethods = ['OLS_gsn', 'OLS_conc', 'W']\nfor policy in policies:\n fig, axes = plt.subplots(2, K, figsize=(10, 8), sharey=True, sharex=True)\n for k in range(K):\n for m in range(len(methods)):\n method = methods[m]\n axes[0, k].errorbar(100*confidence_levels,\n 100*np.mean([lower_coverage[r][policy][method][k,:] for r in xrange(R)],axis=0),\n label = method, \n marker=markers[method],\n color=colors[method], \n linestyle='')\n #print np.mean([lower_coverage[r][policy]['W'][k,:] for r in xrange(R)],axis=0)\n axes[0,k].plot(100*confidence_levels, 100*confidence_levels, color=colors['Nominal'], label='Nominal')\n #axes[0, k].set(adjustable='box-forced', aspect='equal')\n axes[0, k].set_title(\"Lower: beta\"+str(k+1), fontsize = title_font_size)\n axes[0, k].set_ylim([86, 102])\n for method in methods:\n\n axes[1, k].errorbar(100*confidence_levels,\n 100*np.mean([upper_coverage[r][policy][method][k,:] for r in xrange(R)],axis=0),\n label = method,\n marker = markers[method],\n color=colors[method], \n linestyle = '')\n axes[1,k].plot(100*confidence_levels, 100*confidence_levels, color=colors['Nominal'], label='Nominal')\n #axes[1,k].set(adjustable='box-forced', aspect='equal')\n axes[1,k].set_title(\"Upper: beta\"+str(k+1), fontsize = title_font_size)\n # fig.tight_layout()\n\n plt.figlegend( axes[1,0].get_lines(), methods+['Nom'], \n loc = (0.1, 0.01), ncol=5, \n labelspacing=0. , \n fontsize = legend_font_size)\n fig.suptitle(policy, fontsize = title_font_size, fontweight='bold')\n fig.savefig(path+'mab_{}_coverage'.format(policy))\n plt.show()",
"_____no_output_____"
],
[
"#\n# Display coverage results for sum reward\n#\n## Select coverage array from {\"coverage\", \"lower_coverage\", \"upper_coverage\"}\n#coverage_type = \"lower_coverage\"\n#coverage_arr = locals()[coverage_type]\n# For each policy and method, display coverage as a function of confidence level\n\nmethods = ['OLS_gsn', 'OLS_conc', 'W']\nfor policy in policies:\n fig, axes = plt.subplots(ncols=2, figsize=(11, 4), sharey=True, sharex=True)\n for m in range(len(methods)):\n method = methods[m]\n axes[0].errorbar(100*confidence_levels,\n 100*np.mean([lower_sum_coverage[r][policy][method] for r in xrange(R)],axis=0),\n yerr=100*np.std([lower_sum_coverage[r][policy][method] for r in xrange(R)],axis=0)/np.sqrt(R), \n label = method, \n marker=markers[method],\n color=colors[method], \n linestyle='')\n #print np.mean([lower_coverage[r][policy]['W'][k,:] for r in xrange(R)],axis=0)\n axes[0].plot(100*confidence_levels, 100*confidence_levels, color=colors['Nominal'], label='Nominal')\n #axes[0, k].set(adjustable='box-forced', aspect='equal')\n axes[0].set_title(\"Lower: avg reward\", fontsize = title_font_size)\n axes[0].set_ylim([85, 101])\n for method in methods:\n\n axes[1].errorbar(100*confidence_levels,\n 100*np.mean([upper_sum_coverage[r][policy][method] for r in xrange(R)],axis=0),\n yerr= 100*np.std([upper_sum_coverage[r][policy][method] for r in xrange(R)],axis=0)/np.sqrt(R),\n label = method,\n marker = markers[method],\n color=colors[method], \n linestyle = '')\n axes[1].plot(100*confidence_levels, 100*confidence_levels, color=colors['Nominal'], label='Nominal')\n #axes[1,k].set(adjustable='box-forced', aspect='equal')\n axes[1].set_title(\"Upper: avg reward\", fontsize = title_font_size)\n \n # fig.tight_layout()\n handles = axes[1].get_lines() \n axes[1].legend( handles[0:3] + [handles[4]], \n ['OLS_gsn','Nom', 'OLS_conc', 'W'],\n loc='lower right',\n bbox_to_anchor= (1, 0.0), \n ncol=1, \n labelspacing=0. , \n fontsize = legend_font_size)\n fig.suptitle(policy, fontsize = title_font_size, fontweight='bold')\n fig.savefig(path+'mab_sum_{}_coverage'.format(policy))\n plt.show()",
"_____no_output_____"
],
[
"#\n# Display width results\n#\n# For each policy and method, display mean width as a function of confidence level\npolicies = [\"ECB\", \"TS\", 'UCB']\nmethods = ['OLS_gsn', 'OLS_conc', 'W']\nfor policy in policies:\n fig, axes = plt.subplots(1, K, sharey=True)\n for k in range(K):\n \n for method in methods:\n axes[k].errorbar(100*confidence_levels, \\\n np.mean([thresholds_os[r][policy][method][k,:] for r in xrange(R)],axis=0), \\\n np.std([thresholds_os[r][policy][method][k,:] for r in xrange(R)], axis=0),\\\n label = method,\n marker = markers[method],\n color=colors[method],\n linestyle='')\n \n # axes[k].legend(loc='')\n axes[k].set_title('arm_{}'.format(k), fontsize = title_font_size)\n # axes[k].set_yscale('log', nonposy='clip')\n fig.suptitle(policy, fontsize = title_font_size, x=0.5, y=1.05, fontweight='bold')\n# plt.figlegend( axes[0].get_lines(), methods, \n# loc=(0.85, 0.5),\n# ncol = 1,\n# # loc= (0.75, 0.3),\n# labelspacing=0. , \n# fontsize = legend_font_size)\n axes[0].legend( axes[0].get_lines(), \n methods, \n loc = 'upper left', \n ncol=1, \n labelspacing=0. ,\n bbox_to_anchor=(0, 1),\n fontsize = legend_font_size)\n fig.set_size_inches(11, 4, forward=True)\n# fig.savefig(path+'mab_{}_width'.format(policy), bbox_inches='tight', pad_inches=0.1)\n plt.show()\n\n",
"_____no_output_____"
],
[
"#\n# Display width results for avg reward\n#\n# For each policy and method, display mean width as a function of confidence level\npolicies = [\"ECB\", \"TS\", 'UCB']\nmethods = ['OLS_gsn', 'OLS_conc', 'W']\nfor policy in policies:\n fig, axes = plt.subplots()\n \n for method in methods:\n sqwidths = np.array([thresholds_os[r][policy][method]**2 for r in xrange(R)])\n widths = np.sqrt(sqwidths.sum(axis = 1))/2\n axes.errorbar(100*confidence_levels, \\\n np.mean(widths, axis=0), \\\n np.std(widths, axis=0),\\\n label = method,\n marker = markers[method],\n color=colors[method],\n linestyle='')\n \n # axes[k].legend(loc='')\n # axes[k].set_title('arm_{}'.format(k), fontsize = title_font_size)\n # axes[k].set_yscale('log', nonposy='clip')\n fig.suptitle(policy, fontsize = title_font_size, x=0.5, y=1.05, fontweight='bold')\n axes.legend(methods,\n loc='upper left',\n bbox_to_anchor=(0,1),\n fontsize=legend_font_size)\n# plt.figlegend( axes[0].get_lines(), methods, \n# loc=(0.85, 0.5),\n# ncol = 1,\n# # loc= (0.75, 0.3),\n# labelspacing=0. , \n# fontsize = legend_font_size)\n# plt.figlegend( axes.get_lines(), methods, \n# loc = (0.21, -0.01), ncol=1, \n# labelspacing=0. , \n# fontsize = legend_font_size)\n fig.set_size_inches(5.5, 4, forward=True)\n fig.savefig(path+'mab_sum_{}_width'.format(policy), bbox_inches='tight', pad_inches=0.1)\n plt.show()\n\n",
"_____no_output_____"
],
[
"#\n# Visualize distribution of parameter estimation error\n#\n\npolicies = [\"UCB\", 'TS', 'ECB']\nmethods = [\"OLS_gsn\", \"W\"]\n#Plot histograms of errors\n#for policy in policies:\n# fig, axes = plt.subplots(nrows=len(methods), ncols=K, sharex=True)\n# for m in range(len(methods)):\n# method = methods[m]\n# for k in range(K):\n# errors = [normalized_errors[r][policy][method][k] for r in xrange(R)]\n# sns.distplot(errors, \n# kde=False,\n# bins=10,\n# fit = stats.norm, \n# ax=axes[k, m])\n# #axes[k,m].hist([estimates[r][policy][method][k] - reward_means[k] for r in xrange(R)],\n# #bins=50, facecolor = 'g')\n# if k == 0:\n# axes[k,m].set_title(method)\n# fig.suptitle(policy)\n# fig.savefig(path+'mab_{}_histogram'.format(policy))\n# plt.show()\n\n# Plot qqplots of errors\nfor policy in policies:\n fig, axes = plt.subplots(nrows=len(methods), ncols=K, \n sharex=True, sharey=False, \n figsize=(10, 8))\n for m in range(len(methods)):\n method = methods[m]\n for k in range(K):\n errors = [normalized_errors[r][policy][method][k] for r in xrange(R)]\n # sm.graphics.qqplot(errors, line='s', ax=axes[k, m])\n orderedstats, fitparams = stats.probplot(errors, \n dist=\"norm\", plot=None)\n \n axes[k, m].plot(orderedstats[0], orderedstats[1], \n marker='o', markersize=4, \n linestyle='', \n color=colors[method])\n axes[k, m].plot(orderedstats[0], fitparams[0]*orderedstats[0] + fitparams[1], color = colors['Nominal'])\n #axes[k, m].plot(orderedstats[0], fitparams[0]*orderedstats[0] + fitparams[1]) #replot to get orange color\n \n if k == 0:\n axes[k,m].set_title(method, fontsize=title_font_size)\n axes[k,m].set_xlabel(\"\")\n else:\n axes[k,m].set_title(\"\")\n # Display empirical kurtosis to 3 significant figures\n axes[k,m].legend(loc='upper left', \n labels=['Ex.Kurt.: {0:.2g}'.format(\n stats.kurtosis(errors, fisher=True))], fontsize=12)\n fig.suptitle(policy, fontsize=title_font_size, fontweight='bold')\n #fig.set_size_inches(6, 4.5)\n fig.savefig(path+'mab_{}_qq'.format(policy))\n plt.show()\n",
"_____no_output_____"
],
[
"## plot PP Plots for arm\npolicies = [\"UCB\", 'TS', 'ECB']\nmethods = [\"OLS_gsn\", \"W\"]\n\nprobvals = np.linspace(0, 1.0, 101)\nbins = stats.norm.ppf(probvals)\nnormdata = np.random.randn(R)\nfor policy in policies:\n fig, axes = plt.subplots(nrows=len(methods), ncols=K, \n sharex=True, sharey=True, \n figsize=(11, 8))\n for m in range(len(methods)):\n method = methods[m]\n for k in range(K):\n errors = [normalized_errors[r][policy][method][k] for r in xrange(R)]\n datacounts, bins = np.histogram(errors, bins, density=True)\n normcounts, bins = np.histogram(normdata, bins, density=True)\n cumdata = np.cumsum(datacounts)\n cumdata = cumdata/max(cumdata)\n cumnorm = np.cumsum(normcounts)\n cumnorm= cumnorm/max(cumnorm)\n axes[k, m].plot(cumnorm, cumdata, \n marker='o', markersize = 4,\n color = colors[method], \n linestyle=''\n )\n axes[k, m].plot(probvals, probvals, color = colors['Nominal'])\n #axes[k, m].plot(orderedstats[0], fitparams[0]*orderedstats[0] + fitparams[1]) #replot to get orange color\n \n if k == 0:\n axes[k,m].set_title(method, fontsize=title_font_size)\n axes[k,m].set_xlabel(\"\")\n else:\n axes[k,m].set_title(\"\")\n # Display empirical kurtosis to 3 significant figures\n axes[k,m].legend(loc='upper left', \n labels=['Skew: {0:.2g}'.format(\n stats.skew(errors))], fontsize=12)\n fig.suptitle(policy, fontsize=title_font_size, fontweight='bold')\n #fig.set_size_inches(6, 4.5)\n fig.savefig(path+'mab_{}_pp'.format(policy))\n plt.show()\n",
"_____no_output_____"
],
[
"# plot qq plots for arm sums\n\npolicies = [\"UCB\", 'TS', 'ECB']\nmethods = [\"OLS_gsn\", \"W\"]\n\n\n\nfor policy in policies:\n fig, axes = plt.subplots(ncols=len(methods), \n sharex=True, sharey=False, \n figsize=(10, 4))\n for m in range(len(methods)):\n method = methods[m]\n \n errors = [sum_norm[r][policy][method] for r in xrange(R)]\n # sm.graphics.qqplot(errors, line='s', ax=axes[k, m])\n orderedstats, fitparams = stats.probplot(errors, \n dist=\"norm\", plot=None)\n \n axes[m].plot(orderedstats[0], orderedstats[1], \n marker='o', markersize=2, \n linestyle='', \n color=colors[method])\n axes[m].plot(orderedstats[0], fitparams[0]*orderedstats[0] + fitparams[1], color = colors['Nominal'])\n #axes[k, m].plot(orderedstats[0], fitparams[0]*orderedstats[0] + fitparams[1]) #replot to get orange color\n \n \n axes[m].set_title(method, fontsize=title_font_size)\n axes[m].set_xlabel(\"\")\n \n axes[m].set_title(\"\")\n # Display empirical kurtosis to 3 significant figures\n# axes[k,m].legend(loc='upper left', \n# labels=['Ex.Kurt.: {0:.2g}'.format(\n# stats.kurtosis(errors, fisher=True))], fontsize=12)\n fig.suptitle(policy, fontsize=title_font_size, fontweight='bold')\n #fig.set_size_inches(6, 4.5)\n fig.savefig(path+'mab_sum_{}_qq'.format(policy))\n plt.show()\n\n\n\n\n",
"_____no_output_____"
],
[
"# plot pp plots for the sums\npolicies = [\"UCB\", 'TS', 'ECB']\nmethods = [\"OLS_gsn\", \"W\"]\n\nprobvals = np.linspace(0, 1.0, 101)\nzscores = stats.norm.ppf(probvals)\nzscores_arr = np.outer(zscores, np.ones(R))\nbins = stats.norm.ppf(probvals)\nnormdata = np.random.randn(R)\n\nfor policy in policies:\n fig, axes = plt.subplots(ncols=len(methods), \n sharex=True, sharey=False, \n figsize=(11, 4))\n for m in range(len(methods)):\n method = methods[m]\n \n errors = [sum_norm[r][policy][method] for r in xrange(R)]\n cumdata = np.mean(errors <= zscores_arr, axis=1)\n # sm.graphics.qqplot(errors, line='s', ax=axes[k, m])\n# datacounts, bins = np.histogram(errors, bins, density=True)\n# normcounts, bins = np.histogram(normdata, bins, density=True)\n# cumdata = np.cumsum(datacounts)\n# cumdata = cumdata/max(cumdata)\n# cumnorm = np.cumsum(normcounts)\n# cumnorm= cumnorm/max(cumnorm)\n axes[m].plot(probvals, cumdata, \n marker='o', markersize = 4,\n color = colors[method], \n linestyle=''\n )\n axes[m].plot(probvals, probvals, color = colors['Nominal'])\n axes[m].set_title(method, fontsize=title_font_size)\n axes[m].set_xlabel(\"\")\n \n axes[m].set_title(\"\")\n # Display empirical kurtosis to 3 significant figures\n# axes[k,m].legend(loc='upper left', \n# labels=['Ex.Kurt.: {0:.2g}'.format(\n# stats.kurtosis(errors, fisher=True))], fontsize=12)\n fig.suptitle(policy, fontsize=title_font_size, fontweight='bold')\n #fig.set_size_inches(6, 4.5)\n fig.savefig(path+'mab_sum_{}_pp'.format(policy))\n plt.show()\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d00381c6c1c2ecbe86e178df3524a3169607b34d | 255,793 | ipynb | Jupyter Notebook | experiments/tl_1v2/cores-oracle.run1.limited/trials/29/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | experiments/tl_1v2/cores-oracle.run1.limited/trials/29/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | experiments/tl_1v2/cores-oracle.run1.limited/trials/29/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | 86.974838 | 76,160 | 0.75486 | [
[
[
"# Transfer Learning Template",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n%matplotlib inline\n\n \nimport os, json, sys, time, random\nimport numpy as np\nimport torch\nfrom torch.optim import Adam\nfrom easydict import EasyDict\nimport matplotlib.pyplot as plt\n\nfrom steves_models.steves_ptn import Steves_Prototypical_Network\n\nfrom steves_utils.lazy_iterable_wrapper import Lazy_Iterable_Wrapper\nfrom steves_utils.iterable_aggregator import Iterable_Aggregator\nfrom steves_utils.ptn_train_eval_test_jig import PTN_Train_Eval_Test_Jig\nfrom steves_utils.torch_sequential_builder import build_sequential\nfrom steves_utils.torch_utils import get_dataset_metrics, ptn_confusion_by_domain_over_dataloader\nfrom steves_utils.utils_v2 import (per_domain_accuracy_from_confusion, get_datasets_base_path)\nfrom steves_utils.PTN.utils import independent_accuracy_assesment\n\nfrom torch.utils.data import DataLoader\n\nfrom steves_utils.stratified_dataset.episodic_accessor import Episodic_Accessor_Factory\n\nfrom steves_utils.ptn_do_report import (\n get_loss_curve,\n get_results_table,\n get_parameters_table,\n get_domain_accuracies,\n)\n\nfrom steves_utils.transforms import get_chained_transform",
"_____no_output_____"
]
],
[
[
"# Allowed Parameters\nThese are allowed parameters, not defaults\nEach of these values need to be present in the injected parameters (the notebook will raise an exception if they are not present)\n\nPapermill uses the cell tag \"parameters\" to inject the real parameters below this cell.\nEnable tags to see what I mean",
"_____no_output_____"
]
],
[
[
"required_parameters = {\n \"experiment_name\",\n \"lr\",\n \"device\",\n \"seed\",\n \"dataset_seed\",\n \"n_shot\",\n \"n_query\",\n \"n_way\",\n \"train_k_factor\",\n \"val_k_factor\",\n \"test_k_factor\",\n \"n_epoch\",\n \"patience\",\n \"criteria_for_best\",\n \"x_net\",\n \"datasets\",\n \"torch_default_dtype\",\n \"NUM_LOGS_PER_EPOCH\",\n \"BEST_MODEL_PATH\",\n \"x_shape\",\n}",
"_____no_output_____"
],
[
"from steves_utils.CORES.utils import (\n ALL_NODES,\n ALL_NODES_MINIMUM_1000_EXAMPLES,\n ALL_DAYS\n)\n\nfrom steves_utils.ORACLE.utils_v2 import (\n ALL_DISTANCES_FEET_NARROWED,\n ALL_RUNS,\n ALL_SERIAL_NUMBERS,\n)\n\nstandalone_parameters = {}\nstandalone_parameters[\"experiment_name\"] = \"STANDALONE PTN\"\nstandalone_parameters[\"lr\"] = 0.001\nstandalone_parameters[\"device\"] = \"cuda\"\n\nstandalone_parameters[\"seed\"] = 1337\nstandalone_parameters[\"dataset_seed\"] = 1337\n\nstandalone_parameters[\"n_way\"] = 8\nstandalone_parameters[\"n_shot\"] = 3\nstandalone_parameters[\"n_query\"] = 2\nstandalone_parameters[\"train_k_factor\"] = 1\nstandalone_parameters[\"val_k_factor\"] = 2\nstandalone_parameters[\"test_k_factor\"] = 2\n\n\nstandalone_parameters[\"n_epoch\"] = 50\n\nstandalone_parameters[\"patience\"] = 10\nstandalone_parameters[\"criteria_for_best\"] = \"source_loss\"\n\nstandalone_parameters[\"datasets\"] = [\n {\n \"labels\": ALL_SERIAL_NUMBERS,\n \"domains\": ALL_DISTANCES_FEET_NARROWED,\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": os.path.join(get_datasets_base_path(), \"oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl\"),\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [\"unit_mag\", \"minus_two\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE_\"\n },\n {\n \"labels\": ALL_NODES,\n \"domains\": ALL_DAYS,\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": os.path.join(get_datasets_base_path(), \"cores.stratified_ds.2022A.pkl\"),\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [\"unit_power\", \"times_zero\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"CORES_\"\n } \n]\n\nstandalone_parameters[\"torch_default_dtype\"] = \"torch.float32\" \n\n\n\nstandalone_parameters[\"x_net\"] = [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\":[-1, 1, 2, 256]}},\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":1, \"out_channels\":256, \"kernel_size\":(1,7), \"bias\":False, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":256, \"out_channels\":80, \"kernel_size\":(2,7), \"bias\":True, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 80*256, \"out_features\": 256}}, # 80 units per IQ pair\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n]\n\n# Parameters relevant to results\n# These parameters will basically never need to change\nstandalone_parameters[\"NUM_LOGS_PER_EPOCH\"] = 10\nstandalone_parameters[\"BEST_MODEL_PATH\"] = \"./best_model.pth\"\n\n\n\n\n",
"_____no_output_____"
],
[
"# Parameters\nparameters = {\n \"experiment_name\": \"tl_1v2:cores-oracle.run1.limited\",\n \"device\": \"cuda\",\n \"lr\": 0.0001,\n \"n_shot\": 3,\n \"n_query\": 2,\n \"train_k_factor\": 3,\n \"val_k_factor\": 2,\n \"test_k_factor\": 2,\n \"torch_default_dtype\": \"torch.float32\",\n \"n_epoch\": 50,\n \"patience\": 3,\n \"criteria_for_best\": \"target_accuracy\",\n \"x_net\": [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\": [-1, 1, 2, 256]}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 1,\n \"out_channels\": 256,\n \"kernel_size\": [1, 7],\n \"bias\": False,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 256}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 256,\n \"out_channels\": 80,\n \"kernel_size\": [2, 7],\n \"bias\": True,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 20480, \"out_features\": 256}},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\": 256}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n ],\n \"NUM_LOGS_PER_EPOCH\": 10,\n \"BEST_MODEL_PATH\": \"./best_model.pth\",\n \"n_way\": 16,\n \"datasets\": [\n {\n \"labels\": [\n \"1-10.\",\n \"1-11.\",\n \"1-15.\",\n \"1-16.\",\n \"1-17.\",\n \"1-18.\",\n \"1-19.\",\n \"10-4.\",\n \"10-7.\",\n \"11-1.\",\n \"11-14.\",\n \"11-17.\",\n \"11-20.\",\n \"11-7.\",\n \"13-20.\",\n \"13-8.\",\n \"14-10.\",\n \"14-11.\",\n \"14-14.\",\n \"14-7.\",\n \"15-1.\",\n \"15-20.\",\n \"16-1.\",\n \"16-16.\",\n \"17-10.\",\n \"17-11.\",\n \"17-2.\",\n \"19-1.\",\n \"19-16.\",\n \"19-19.\",\n \"19-20.\",\n \"19-3.\",\n \"2-10.\",\n \"2-11.\",\n \"2-17.\",\n \"2-18.\",\n \"2-20.\",\n \"2-3.\",\n \"2-4.\",\n \"2-5.\",\n \"2-6.\",\n \"2-7.\",\n \"2-8.\",\n \"3-13.\",\n \"3-18.\",\n \"3-3.\",\n \"4-1.\",\n \"4-10.\",\n \"4-11.\",\n \"4-19.\",\n \"5-5.\",\n \"6-15.\",\n \"7-10.\",\n \"7-14.\",\n \"8-18.\",\n \"8-20.\",\n \"8-3.\",\n \"8-8.\",\n ],\n \"domains\": [1, 2, 3, 4, 5],\n \"num_examples_per_domain_per_label\": -1,\n \"pickle_path\": \"/root/csc500-main/datasets/cores.stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [],\n \"episode_transforms\": [],\n \"domain_prefix\": \"CORES_\",\n },\n {\n \"labels\": [\n \"3123D52\",\n \"3123D65\",\n \"3123D79\",\n \"3123D80\",\n \"3123D54\",\n \"3123D70\",\n \"3123D7B\",\n \"3123D89\",\n \"3123D58\",\n \"3123D76\",\n \"3123D7D\",\n \"3123EFE\",\n \"3123D64\",\n \"3123D78\",\n \"3123D7E\",\n \"3124E4A\",\n ],\n \"domains\": [32, 38, 8, 44, 14, 50, 20, 26],\n \"num_examples_per_domain_per_label\": 2000,\n \"pickle_path\": \"/root/csc500-main/datasets/oracle.Run1_10kExamples_stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE.run1_\",\n },\n ],\n \"dataset_seed\": 500,\n \"seed\": 500,\n}\n",
"_____no_output_____"
],
[
"# Set this to True if you want to run this template directly\nSTANDALONE = False\nif STANDALONE:\n print(\"parameters not injected, running with standalone_parameters\")\n parameters = standalone_parameters\n\nif not 'parameters' in locals() and not 'parameters' in globals():\n raise Exception(\"Parameter injection failed\")\n\n#Use an easy dict for all the parameters\np = EasyDict(parameters)\n\nif \"x_shape\" not in p:\n p.x_shape = [2,256] # Default to this if we dont supply x_shape\n\n\nsupplied_keys = set(p.keys())\n\nif supplied_keys != required_parameters:\n print(\"Parameters are incorrect\")\n if len(supplied_keys - required_parameters)>0: print(\"Shouldn't have:\", str(supplied_keys - required_parameters))\n if len(required_parameters - supplied_keys)>0: print(\"Need to have:\", str(required_parameters - supplied_keys))\n raise RuntimeError(\"Parameters are incorrect\")",
"_____no_output_____"
],
[
"###################################\n# Set the RNGs and make it all deterministic\n###################################\nnp.random.seed(p.seed)\nrandom.seed(p.seed)\ntorch.manual_seed(p.seed)\n\ntorch.use_deterministic_algorithms(True) ",
"_____no_output_____"
],
[
"###########################################\n# The stratified datasets honor this\n###########################################\ntorch.set_default_dtype(eval(p.torch_default_dtype))",
"_____no_output_____"
],
[
"###################################\n# Build the network(s)\n# Note: It's critical to do this AFTER setting the RNG\n###################################\nx_net = build_sequential(p.x_net)",
"_____no_output_____"
],
[
"start_time_secs = time.time()",
"_____no_output_____"
],
[
"p.domains_source = []\np.domains_target = []\n\n\ntrain_original_source = []\nval_original_source = []\ntest_original_source = []\n\ntrain_original_target = []\nval_original_target = []\ntest_original_target = []",
"_____no_output_____"
],
[
"# global_x_transform_func = lambda x: normalize(x.to(torch.get_default_dtype()), \"unit_power\") # unit_power, unit_mag\n# global_x_transform_func = lambda x: normalize(x, \"unit_power\") # unit_power, unit_mag",
"_____no_output_____"
],
[
"def add_dataset(\n labels,\n domains,\n pickle_path,\n x_transforms,\n episode_transforms,\n domain_prefix,\n num_examples_per_domain_per_label,\n source_or_target_dataset:str,\n iterator_seed=p.seed,\n dataset_seed=p.dataset_seed,\n n_shot=p.n_shot,\n n_way=p.n_way,\n n_query=p.n_query,\n train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),\n):\n \n if x_transforms == []: x_transform = None\n else: x_transform = get_chained_transform(x_transforms)\n \n if episode_transforms == []: episode_transform = None\n else: raise Exception(\"episode_transforms not implemented\")\n \n episode_transform = lambda tup, _prefix=domain_prefix: (_prefix + str(tup[0]), tup[1])\n\n\n eaf = Episodic_Accessor_Factory(\n labels=labels,\n domains=domains,\n num_examples_per_domain_per_label=num_examples_per_domain_per_label,\n iterator_seed=iterator_seed,\n dataset_seed=dataset_seed,\n n_shot=n_shot,\n n_way=n_way,\n n_query=n_query,\n train_val_test_k_factors=train_val_test_k_factors,\n pickle_path=pickle_path,\n x_transform_func=x_transform,\n )\n\n train, val, test = eaf.get_train(), eaf.get_val(), eaf.get_test()\n train = Lazy_Iterable_Wrapper(train, episode_transform)\n val = Lazy_Iterable_Wrapper(val, episode_transform)\n test = Lazy_Iterable_Wrapper(test, episode_transform)\n\n if source_or_target_dataset==\"source\":\n train_original_source.append(train)\n val_original_source.append(val)\n test_original_source.append(test)\n\n p.domains_source.extend(\n [domain_prefix + str(u) for u in domains]\n )\n elif source_or_target_dataset==\"target\":\n train_original_target.append(train)\n val_original_target.append(val)\n test_original_target.append(test)\n p.domains_target.extend(\n [domain_prefix + str(u) for u in domains]\n )\n else:\n raise Exception(f\"invalid source_or_target_dataset: {source_or_target_dataset}\")\n ",
"_____no_output_____"
],
[
"for ds in p.datasets:\n add_dataset(**ds)",
"_____no_output_____"
],
[
"# from steves_utils.CORES.utils import (\n# ALL_NODES,\n# ALL_NODES_MINIMUM_1000_EXAMPLES,\n# ALL_DAYS\n# )\n\n# add_dataset(\n# labels=ALL_NODES,\n# domains = ALL_DAYS,\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"cores.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"cores_{u}\"\n# )",
"_____no_output_____"
],
[
"# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n\n\n# add_dataset(\n# labels=ALL_SERIAL_NUMBERS,\n# domains = list(set(ALL_DISTANCES_FEET) - {2,62}),\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"source\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"oracle1_{u}\"\n# )\n",
"_____no_output_____"
],
[
"# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n\n\n# add_dataset(\n# labels=ALL_SERIAL_NUMBERS,\n# domains = list(set(ALL_DISTANCES_FEET) - {2,62,56}),\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"source\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"oracle2_{u}\"\n# )",
"_____no_output_____"
],
[
"# add_dataset(\n# labels=list(range(19)),\n# domains = [0,1,2],\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"metehan.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"met_{u}\"\n# )",
"_____no_output_____"
],
[
"# # from steves_utils.wisig.utils import (\n# # ALL_NODES_MINIMUM_100_EXAMPLES,\n# # ALL_NODES_MINIMUM_500_EXAMPLES,\n# # ALL_NODES_MINIMUM_1000_EXAMPLES,\n# # ALL_DAYS\n# # )\n\n# import steves_utils.wisig.utils as wisig\n\n\n# add_dataset(\n# labels=wisig.ALL_NODES_MINIMUM_100_EXAMPLES,\n# domains = wisig.ALL_DAYS,\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"wisig.node3-19.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"wisig_{u}\"\n# )",
"_____no_output_____"
],
[
"###################################\n# Build the dataset\n###################################\ntrain_original_source = Iterable_Aggregator(train_original_source, p.seed)\nval_original_source = Iterable_Aggregator(val_original_source, p.seed)\ntest_original_source = Iterable_Aggregator(test_original_source, p.seed)\n\n\ntrain_original_target = Iterable_Aggregator(train_original_target, p.seed)\nval_original_target = Iterable_Aggregator(val_original_target, p.seed)\ntest_original_target = Iterable_Aggregator(test_original_target, p.seed)\n\n# For CNN We only use X and Y. And we only train on the source.\n# Properly form the data using a transform lambda and Lazy_Iterable_Wrapper. Finally wrap them in a dataloader\n\ntransform_lambda = lambda ex: ex[1] # Original is (<domain>, <episode>) so we strip down to episode only\n\ntrain_processed_source = Lazy_Iterable_Wrapper(train_original_source, transform_lambda)\nval_processed_source = Lazy_Iterable_Wrapper(val_original_source, transform_lambda)\ntest_processed_source = Lazy_Iterable_Wrapper(test_original_source, transform_lambda)\n\ntrain_processed_target = Lazy_Iterable_Wrapper(train_original_target, transform_lambda)\nval_processed_target = Lazy_Iterable_Wrapper(val_original_target, transform_lambda)\ntest_processed_target = Lazy_Iterable_Wrapper(test_original_target, transform_lambda)\n\ndatasets = EasyDict({\n \"source\": {\n \"original\": {\"train\":train_original_source, \"val\":val_original_source, \"test\":test_original_source},\n \"processed\": {\"train\":train_processed_source, \"val\":val_processed_source, \"test\":test_processed_source}\n },\n \"target\": {\n \"original\": {\"train\":train_original_target, \"val\":val_original_target, \"test\":test_original_target},\n \"processed\": {\"train\":train_processed_target, \"val\":val_processed_target, \"test\":test_processed_target}\n },\n})",
"_____no_output_____"
],
[
"from steves_utils.transforms import get_average_magnitude, get_average_power\n\nprint(set([u for u,_ in val_original_source]))\nprint(set([u for u,_ in val_original_target]))\n\ns_x, s_y, q_x, q_y, _ = next(iter(train_processed_source))\nprint(s_x)\n\n# for ds in [\n# train_processed_source,\n# val_processed_source,\n# test_processed_source,\n# train_processed_target,\n# val_processed_target,\n# test_processed_target\n# ]:\n# for s_x, s_y, q_x, q_y, _ in ds:\n# for X in (s_x, q_x):\n# for x in X:\n# assert np.isclose(get_average_magnitude(x.numpy()), 1.0)\n# assert np.isclose(get_average_power(x.numpy()), 1.0)\n ",
"{'ORACLE.run1_26', 'ORACLE.run1_50', 'ORACLE.run1_20', 'ORACLE.run1_8', 'ORACLE.run1_14', 'ORACLE.run1_32', 'ORACLE.run1_44', 'ORACLE.run1_38'}\n"
],
[
"###################################\n# Build the model\n###################################\n# easfsl only wants a tuple for the shape\nmodel = Steves_Prototypical_Network(x_net, device=p.device, x_shape=tuple(p.x_shape))\noptimizer = Adam(params=model.parameters(), lr=p.lr)",
"(2, 256)\n"
],
[
"###################################\n# train\n###################################\njig = PTN_Train_Eval_Test_Jig(model, p.BEST_MODEL_PATH, p.device)\n\njig.train(\n train_iterable=datasets.source.processed.train,\n source_val_iterable=datasets.source.processed.val,\n target_val_iterable=datasets.target.processed.val,\n num_epochs=p.n_epoch,\n num_logs_per_epoch=p.NUM_LOGS_PER_EPOCH,\n patience=p.patience,\n optimizer=optimizer,\n criteria_for_best=p.criteria_for_best,\n)",
"epoch: 1, [batch: 1 / 6720], examples_per_second: 32.6377, train_label_loss: 2.7210, \n"
],
[
"total_experiment_time_secs = time.time() - start_time_secs",
"_____no_output_____"
],
[
"###################################\n# Evaluate the model\n###################################\nsource_test_label_accuracy, source_test_label_loss = jig.test(datasets.source.processed.test)\ntarget_test_label_accuracy, target_test_label_loss = jig.test(datasets.target.processed.test)\n\nsource_val_label_accuracy, source_val_label_loss = jig.test(datasets.source.processed.val)\ntarget_val_label_accuracy, target_val_label_loss = jig.test(datasets.target.processed.val)\n\nhistory = jig.get_history()\n\ntotal_epochs_trained = len(history[\"epoch_indices\"])\n\nval_dl = Iterable_Aggregator((datasets.source.original.val,datasets.target.original.val))\n\nconfusion = ptn_confusion_by_domain_over_dataloader(model, p.device, val_dl)\nper_domain_accuracy = per_domain_accuracy_from_confusion(confusion)\n\n# Add a key to per_domain_accuracy for if it was a source domain\nfor domain, accuracy in per_domain_accuracy.items():\n per_domain_accuracy[domain] = {\n \"accuracy\": accuracy,\n \"source?\": domain in p.domains_source\n }\n\n# Do an independent accuracy assesment JUST TO BE SURE!\n# _source_test_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.test, p.device)\n# _target_test_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.test, p.device)\n# _source_val_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.val, p.device)\n# _target_val_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.val, p.device)\n\n# assert(_source_test_label_accuracy == source_test_label_accuracy)\n# assert(_target_test_label_accuracy == target_test_label_accuracy)\n# assert(_source_val_label_accuracy == source_val_label_accuracy)\n# assert(_target_val_label_accuracy == target_val_label_accuracy)\n\nexperiment = {\n \"experiment_name\": p.experiment_name,\n \"parameters\": dict(p),\n \"results\": {\n \"source_test_label_accuracy\": source_test_label_accuracy,\n \"source_test_label_loss\": source_test_label_loss,\n \"target_test_label_accuracy\": target_test_label_accuracy,\n \"target_test_label_loss\": target_test_label_loss,\n \"source_val_label_accuracy\": source_val_label_accuracy,\n \"source_val_label_loss\": source_val_label_loss,\n \"target_val_label_accuracy\": target_val_label_accuracy,\n \"target_val_label_loss\": target_val_label_loss,\n \"total_epochs_trained\": total_epochs_trained,\n \"total_experiment_time_secs\": total_experiment_time_secs,\n \"confusion\": confusion,\n \"per_domain_accuracy\": per_domain_accuracy,\n },\n \"history\": history,\n \"dataset_metrics\": get_dataset_metrics(datasets, \"ptn\"),\n}",
"_____no_output_____"
],
[
"ax = get_loss_curve(experiment)\nplt.show()",
"_____no_output_____"
],
[
"get_results_table(experiment)",
"_____no_output_____"
],
[
"get_domain_accuracies(experiment)",
"_____no_output_____"
],
[
"print(\"Source Test Label Accuracy:\", experiment[\"results\"][\"source_test_label_accuracy\"], \"Target Test Label Accuracy:\", experiment[\"results\"][\"target_test_label_accuracy\"])\nprint(\"Source Val Label Accuracy:\", experiment[\"results\"][\"source_val_label_accuracy\"], \"Target Val Label Accuracy:\", experiment[\"results\"][\"target_val_label_accuracy\"])",
"Source Test Label Accuracy: 0.7997721354166667 Target Test Label Accuracy: 0.9880266853932584\nSource Val Label Accuracy: 0.7990559895833333 Target Val Label Accuracy: 0.9888214285714285\n"
],
[
"json.dumps(experiment)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d003923170097cca0d4cfccfc3332ba0ba18f4e7 | 5,085 | ipynb | Jupyter Notebook | samples/convert_old_input_card.ipynb | kidddddd1984/CVM | 676f7f4df188742cd739a6cd82e9236afecc3b94 | [
"BSD-3-Clause"
] | null | null | null | samples/convert_old_input_card.ipynb | kidddddd1984/CVM | 676f7f4df188742cd739a6cd82e9236afecc3b94 | [
"BSD-3-Clause"
] | 1 | 2015-08-23T13:08:35.000Z | 2017-04-07T09:27:40.000Z | samples/convert_old_input_card.ipynb | kidddddd1984/CVM | 676f7f4df188742cd739a6cd82e9236afecc3b94 | [
"BSD-3-Clause"
] | null | null | null | 23.761682 | 146 | 0.466077 | [
[
[
"# Convert old input card\n\n1. meta and experiment",
"_____no_output_____"
]
],
[
[
"from ruamel.yaml import YAML\nfrom cvm.utils import get_inp\nimport sys\n\nyaml = YAML()\nyaml.indent(mapping=4, sequence=4, offset=2)\nyaml.default_flow_style = None\nyaml.width = 120",
"_____no_output_____"
],
[
"inp = get_inp('<old_input_card.json>')",
"_____no_output_____"
],
[
"meta = dict(host=inp['host'], impurity=inp['impurity'], prefix=inp['prefix'], description=inp['description'], structure=inp['structure'])\nexperiment = dict(temperature=inp['experiment'][0]['temp'], concentration=inp['experiment'][0]['c'])\n\ntmp = {'meta': meta, 'experiment': experiment}",
"_____no_output_____"
],
[
"tmp",
"_____no_output_____"
],
[
"with open('input.yml', 'w') as f:\n yaml.dump(tmp, f)",
"_____no_output_____"
]
],
[
[
"2. enegires",
"_____no_output_____"
]
],
[
[
"def extractor(s, prefix):\n print(s['label'])\n print(s['transfer'])\n print(s['temp'])\n data = s['datas']\n lattice = data['lattice_c']\n host=data['host_en']\n \n n_ens = {}\n for i in range(11):\n s_i = str(i + 1)\n l = 'pair' + s_i\n n_ens[s_i + '_II'] = data[l][0]['energy']\n n_ens[s_i + '_IH'] = data[l][1]['energy']\n n_ens[s_i + '_HH'] = data[l][2]['energy']\n \n normalizer = dict(lattice=lattice, **n_ens)\n clusters = dict(\n lattice=lattice,\n host=host,\n Rh4=data['tetra'][0]['energy'],\n Rh3Pd1=data['tetra'][1]['energy'],\n Rh2Pd2=data['tetra'][2]['energy'],\n Rh1Pd3=data['tetra'][3]['energy'],\n Pd4=data['tetra'][4]['energy'],\n )\n \n n_name = prefix + '_normalizer.csv'\n c_name = prefix + '_clusters.csv'\n \n print(n_name)\n print(c_name)\n print()\n \n pd.DataFrame(normalizer).to_csv(n_name, index=False)\n pd.DataFrame(clusters).to_csv(c_name, index=False)\n ",
"_____no_output_____"
],
[
"for i, s in enumerate(inp['series']):\n extractor(s, str(i))",
"$T_\\mathrm{FD}=800$K\n[[1, 11, 2]]\n[400, 1290, 50]\n0_normalizer.csv\n0_clusters.csv\n\n$T_\\mathrm{FD}=1000$K\n[[1, 11, 2]]\n[400, 1550, 50]\n1_normalizer.csv\n1_clusters.csv\n\n$T_\\mathrm{FD}=1200$K\n[[1, 11, 2]]\n[400, 1700, 50]\n2_normalizer.csv\n2_clusters.csv\n\n$T_\\mathrm{FD}=1400$K\n[[1, 11, 2]]\n[500, 1700, 50]\n3_normalizer.csv\n3_clusters.csv\n\n$T_\\mathrm{FD}=1600$K\n[[1, 11, 2]]\n[500, 1870, 50]\n4_normalizer.csv\n4_clusters.csv\n\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d003a30e73b102910e1378689f851e9ea046f464 | 8,294 | ipynb | Jupyter Notebook | simple_generative_adversarial_net/MNIST_GANs.ipynb | s-mostafa-a/a | dfaa84cb6a09f8d0dafc03a438f02779df83b34d | [
"MIT"
] | 5 | 2020-06-22T13:04:21.000Z | 2022-01-14T01:01:47.000Z | simple_generative_adversarial_net/MNIST_GANs.ipynb | s-mostafa-a/a | dfaa84cb6a09f8d0dafc03a438f02779df83b34d | [
"MIT"
] | 8 | 2021-06-08T21:39:00.000Z | 2022-02-15T00:05:04.000Z | simple_generative_adversarial_net/MNIST_GANs.ipynb | s-mostafa-a/a | dfaa84cb6a09f8d0dafc03a438f02779df83b34d | [
"MIT"
] | 1 | 2021-02-07T23:36:54.000Z | 2021-02-07T23:36:54.000Z | 44.117021 | 271 | 0.487943 | [
[
[
"<a href=\"https://colab.research.google.com/github/s-mostafa-a/pytorch_learning/blob/master/simple_generative_adversarial_net/MNIST_GANs.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torchvision.transforms import ToTensor, Normalize, Compose\nfrom torchvision.datasets import MNIST\nimport torch.nn as nn\nfrom torch.utils.data import DataLoader\nfrom torchvision.utils import save_image\nimport os\nclass DeviceDataLoader:\n def __init__(self, dl, device):\n self.dl = dl\n self.device = device\n\n def __iter__(self):\n for b in self.dl:\n yield self.to_device(b, self.device)\n\n def __len__(self):\n return len(self.dl)\n\n def to_device(self, data, device):\n if isinstance(data, (list, tuple)):\n return [self.to_device(x, device) for x in data]\n return data.to(device, non_blocking=True)\n\nclass MNIST_GANS:\n def __init__(self, dataset, image_size, device, num_epochs=50, loss_function=nn.BCELoss(), batch_size=100,\n hidden_size=2561, latent_size=64):\n self.device = device\n bare_data_loader = DataLoader(dataset, batch_size, shuffle=True)\n self.data_loader = DeviceDataLoader(bare_data_loader, device)\n self.loss_function = loss_function\n self.hidden_size = hidden_size\n self.latent_size = latent_size\n self.batch_size = batch_size\n self.D = nn.Sequential(\n nn.Linear(image_size, hidden_size),\n nn.LeakyReLU(0.2),\n nn.Linear(hidden_size, hidden_size),\n nn.LeakyReLU(0.2),\n nn.Linear(hidden_size, 1),\n nn.Sigmoid())\n self.G = nn.Sequential(\n nn.Linear(latent_size, hidden_size),\n nn.ReLU(),\n nn.Linear(hidden_size, hidden_size),\n nn.ReLU(),\n nn.Linear(hidden_size, image_size),\n nn.Tanh())\n self.d_optimizer = torch.optim.Adam(self.D.parameters(), lr=0.0002)\n self.g_optimizer = torch.optim.Adam(self.G.parameters(), lr=0.0002)\n self.sample_dir = './../data/mnist_samples'\n if not os.path.exists(self.sample_dir):\n os.makedirs(self.sample_dir)\n self.G.to(device)\n self.D.to(device)\n self.sample_vectors = torch.randn(self.batch_size, self.latent_size).to(self.device)\n self.num_epochs = num_epochs\n\n @staticmethod\n def denormalize(x):\n out = (x + 1) / 2\n return out.clamp(0, 1)\n\n def reset_grad(self):\n self.d_optimizer.zero_grad()\n self.g_optimizer.zero_grad()\n\n def train_discriminator(self, images):\n real_labels = torch.ones(self.batch_size, 1).to(self.device)\n fake_labels = torch.zeros(self.batch_size, 1).to(self.device)\n\n outputs = self.D(images)\n d_loss_real = self.loss_function(outputs, real_labels)\n real_score = outputs\n\n new_sample_vectors = torch.randn(self.batch_size, self.latent_size).to(self.device)\n fake_images = self.G(new_sample_vectors)\n outputs = self.D(fake_images)\n d_loss_fake = self.loss_function(outputs, fake_labels)\n fake_score = outputs\n\n d_loss = d_loss_real + d_loss_fake\n self.reset_grad()\n d_loss.backward()\n self.d_optimizer.step()\n\n return d_loss, real_score, fake_score\n\n def train_generator(self):\n new_sample_vectors = torch.randn(self.batch_size, self.latent_size).to(self.device)\n fake_images = self.G(new_sample_vectors)\n labels = torch.ones(self.batch_size, 1).to(self.device)\n g_loss = self.loss_function(self.D(fake_images), labels)\n\n self.reset_grad()\n g_loss.backward()\n self.g_optimizer.step()\n return g_loss, fake_images\n\n def save_fake_images(self, index):\n fake_images = self.G(self.sample_vectors)\n fake_images = fake_images.reshape(fake_images.size(0), 1, 28, 28)\n fake_fname = 'fake_images-{0:0=4d}.png'.format(index)\n print('Saving', fake_fname)\n save_image(self.denormalize(fake_images), os.path.join(self.sample_dir, fake_fname),\n nrow=10)\n\n def run(self):\n total_step = len(self.data_loader)\n d_losses, g_losses, real_scores, fake_scores = [], [], [], []\n\n for epoch in range(self.num_epochs):\n for i, (images, _) in enumerate(self.data_loader):\n images = images.reshape(self.batch_size, -1)\n\n d_loss, real_score, fake_score = self.train_discriminator(images)\n g_loss, fake_images = self.train_generator()\n\n if (i + 1) % 600 == 0:\n d_losses.append(d_loss.item())\n g_losses.append(g_loss.item())\n real_scores.append(real_score.mean().item())\n fake_scores.append(fake_score.mean().item())\n print(f'''Epoch [{epoch}/{self.num_epochs}], Step [{i + 1}/{\n total_step}], d_loss: {d_loss.item():.4f}, g_loss: {g_loss.item():.4f}, D(x): {\n real_score.mean().item():.2f}, D(G(z)): {fake_score.mean().item():.2f}''')\n self.save_fake_images(epoch + 1)\n\n\ndevice = torch.device('cuda' if torch.cuda.is_available() else 'cpu')\nmnist = MNIST(root='./../data', train=True, download=True, transform=Compose([ToTensor(), Normalize(mean=(0.5,), std=(0.5,))]))\nimage_size = mnist.data[0].flatten().size()[0]\ngans = MNIST_GANS(dataset=mnist, image_size=image_size, device=device)\ngans.run()\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
d003b29dce0beb4d9f782ffacfd931f35fe514a0 | 196,103 | ipynb | Jupyter Notebook | chapter09_numoptim/04_energy.ipynb | aaazzz640/cookbook-2nd-code | c0edeb78fe5a16e64d1210437470b00572211a82 | [
"MIT"
] | 645 | 2018-02-01T09:16:45.000Z | 2022-03-03T17:47:59.000Z | chapter09_numoptim/04_energy.ipynb | aaazzz640/cookbook-2nd-code | c0edeb78fe5a16e64d1210437470b00572211a82 | [
"MIT"
] | 3 | 2019-03-11T09:47:21.000Z | 2022-01-11T06:32:00.000Z | chapter09_numoptim/04_energy.ipynb | aaazzz640/cookbook-2nd-code | c0edeb78fe5a16e64d1210437470b00572211a82 | [
"MIT"
] | 418 | 2018-02-13T03:17:05.000Z | 2022-03-18T21:04:45.000Z | 700.367857 | 140,858 | 0.93791 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d003c7789366a06e031b6b98c9297415273c208b | 30,584 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Statistics-checkpoint.ipynb | IntelligentQuadruped/Vision_Analysis | 95b8a23abd9773e2e62f0849e9ddd81465851ff3 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Statistics-checkpoint.ipynb | IntelligentQuadruped/Vision_Analysis | 95b8a23abd9773e2e62f0849e9ddd81465851ff3 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Statistics-checkpoint.ipynb | IntelligentQuadruped/Vision_Analysis | 95b8a23abd9773e2e62f0849e9ddd81465851ff3 | [
"MIT"
] | null | null | null | 111.214545 | 17,450 | 0.816603 | [
[
[
"import pandas as pd\nimport numpy as np \nimport os\nimport matplotlib.mlab as mlab\nimport matplotlib.pyplot as plt\nimport seaborn\n\ndef filterOutlier(data_list,z_score_threshold=3.5):\n\t\"\"\"\n\tFilters out outliers using the modified Z-Score method.\n\t\"\"\"\n\t# n = len(data_list)\n\t# z_score_threshold = (n-1)/np.sqrt(n)\n\tdata = np.array(data_list)\n\tmedian = np.median(data)\n\tdeviation = np.median([np.abs(x - median) for x in data])\n\tz_scores = [0.675*(x - median)/deviation for x in data]\n\tdata_out = data[np.where(np.abs(z_scores) < z_score_threshold)].tolist()\n\toutput = data_out if len(data_out) > 0 else data_list\n\treturn output\n\ndata_dir = [\"./data/sample_obstacle_course\"]\n# data_dir = ['./windmachine']\n\ndata = []\nfor data_path in data_dir:\n for f in os.listdir(data_path):\n if \"d\" in f:\n try:\n path = os.path.join(data_path,f)\n matrix = np.load(path)\n matrix[matrix > 4000] = 0.0\n nan = len(matrix[matrix < 1])\n total = len(matrix.flatten())\n result = 1 - nan/total\n data.append(result)\n # if True:\n # plt.figure()\n # plt.title(f)\n # plt.imshow(matrix)\n # plt.show()\n except TypeError:\n path = os.path.join(data_path,f)\n d= np.load(path)\n # for i in range(5):\n # s = 'arr_{}'.format(i+1)\n s = 'arr_1'\n matrix = d[s]\n nan = len(matrix[matrix < 1])\n total = len(matrix.flatten())\n result = 1 - nan/total\n data.append(result)\n d.close()\n\n",
"_____no_output_____"
],
[
"# data = filterOutlier(data)\ndata = np.array(data)\ndata = data[abs(data - np.mean(data)) < 3 * np.std(data)].tolist()\nprint(data)",
"[0.3932259114583333, 0.6301888020833333, 0.42250651041666665, 0.39485351562500004, 0.36089843749999995, 0.38500651041666667, 0.40374023437499995, 0.38688151041666663, 0.37723307291666663, 0.37966796875, 0.36004882812500005, 0.3885579427083333, 0.39791341145833337, 0.41855794270833335, 0.37113281249999996, 0.36710611979166663, 0.39001302083333333, 0.380146484375, 0.40511718750000003, 0.3915559895833334, 0.3920475260416667, 0.4053483072916667, 0.4256022135416667, 0.376572265625, 0.36972005208333336, 0.38937825520833336, 0.3711686197916667, 0.3714322916666667, 0.3629752604166666, 0.38073893229166667, 0.39175781249999997, 0.3871028645833333, 0.3907942708333333, 0.3756803385416667, 0.6179752604166666, 0.351708984375, 0.3735677083333333, 0.40713867187499997, 0.3888736979166667, 0.38099283854166666, 0.3637239583333334, 0.3800748697916667, 0.6326595052083333, 0.39737304687499997, 0.389892578125, 0.37627929687499995, 0.40317708333333335, 0.39241210937500004, 0.39200846354166663, 0.35833007812499995, 0.401865234375, 0.3888606770833334, 0.4186393229166666, 0.38731770833333334, 0.39966145833333333, 0.37683268229166667, 0.4180436197916667, 0.37835937500000005, 0.61826171875, 0.359130859375, 0.3953255208333334, 0.4156998697916666, 0.3891829427083333, 0.3800716145833334, 0.4148470052083333, 0.3614127604166667, 0.3944596354166666, 0.35552734374999995, 0.62669921875, 0.36856119791666664, 0.38590820312499996, 0.35775716145833336, 0.37792317708333334, 0.415166015625, 0.38736979166666663, 0.4974446614583333, 0.420791015625, 0.3800325520833333, 0.397841796875, 0.39087565104166666, 0.36547200520833334, 0.3853483072916667, 0.39175455729166664, 0.3910286458333333, 0.37614257812499996, 0.6078743489583334, 0.3724088541666667, 0.37002604166666664, 0.38925130208333336, 0.3859765625, 0.3589388020833333, 0.379658203125, 0.38716796875000004, 0.3906575520833333, 0.35819010416666663, 0.3874934895833333, 0.3830891927083333, 0.37753255208333336, 0.3876529947916667, 0.62359375, 0.42361002604166664, 0.39795572916666666, 0.40796549479166666, 0.3660611979166667, 0.3902213541666667, 0.38036783854166667, 0.3928776041666666, 0.3608528645833333, 0.42780273437500005, 0.36979492187499996, 0.39097981770833334, 0.39128906249999995, 0.3887239583333333, 0.41615885416666665, 0.4128125, 0.39660481770833333, 0.42292968750000004, 0.38151041666666663, 0.36958984375000004, 0.4820735677083333, 0.39451171875, 0.3592903645833333, 0.489599609375, 0.38550130208333333, 0.36537109375, 0.4050162760416667, 0.38827473958333336, 0.3583333333333333, 0.3809635416666667, 0.3656998697916667, 0.39122395833333334, 0.35639648437500004, 0.6321679687499999, 0.6344759114583334, 0.3949674479166667, 0.391572265625, 0.408671875, 0.38786132812499996, 0.35518880208333337, 0.3744498697916666, 0.3823046875, 0.387451171875, 0.3672428385416666, 0.39181966145833336, 0.3573697916666667, 0.4181901041666667, 0.3956477864583333, 0.38620442708333336, 0.3951497395833333, 0.38696940104166666, 0.4101041666666667, 0.38151041666666663, 0.39074869791666667, 0.42233723958333336, 0.3858138020833334, 0.3779850260416666, 0.38117187500000005, 0.46586914062499996, 0.40561848958333335, 0.5841536458333334, 0.5134537760416666, 0.38497395833333337, 0.3867057291666667, 0.35890625, 0.38930338541666665, 0.3563899739583334, 0.38498046875, 0.4090625, 0.3872688802083334, 0.3510970052083333, 0.38242513020833335, 0.40417317708333333, 0.38650065104166664, 0.359638671875, 0.3801497395833333, 0.5955696614583333, 0.5740592447916666, 0.5324251302083334, 0.37850911458333336, 0.36517578125000005, 0.352880859375, 0.4158463541666667, 0.40673502604166667, 0.40416341145833334, 0.46263020833333335, 0.3759440104166667, 0.3709798177083333, 0.383642578125, 0.3900390625, 0.42862304687499997, 0.3873046875, 0.38951497395833334, 0.3670703125, 0.41754882812500005, 0.40291015625000004, 0.4036100260416666, 0.373037109375, 0.3931412760416667, 0.4125325520833333, 0.38467447916666664, 0.38144856770833335, 0.3680501302083333, 0.5898567708333333, 0.364990234375, 0.39597005208333336, 0.3877278645833333, 0.34448893229166666, 0.374013671875, 0.41642252604166663, 0.38013020833333333, 0.3947623697916667, 0.384287109375, 0.37880533854166665, 0.5824967447916667, 0.572744140625, 0.38711588541666664, 0.391259765625, 0.588916015625, 0.550478515625, 0.348037109375, 0.3776985677083333, 0.41786783854166665, 0.36956054687499995, 0.3633040364583333, 0.39689453124999996, 0.39021809895833337, 0.3796614583333333, 0.37215169270833337, 0.3716438802083334, 0.391103515625, 0.4244466145833333, 0.3832389322916666, 0.40748697916666665, 0.3676432291666667, 0.458505859375, 0.38344401041666665, 0.36234374999999996, 0.3531022135416667, 0.54416015625, 0.583681640625, 0.590107421875, 0.40538411458333334, 0.3919010416666666, 0.3497493489583333, 0.3794401041666666, 0.3833235677083333, 0.3605240885416666, 0.3871451822916666, 0.3640559895833333, 0.37371093749999995, 0.4276790364583334, 0.3858040364583334, 0.39063151041666666, 0.39489908854166667, 0.415556640625, 0.40194661458333336, 0.40344401041666667, 0.4569010416666667, 0.4680826822916667, 0.4044791666666666, 0.418427734375, 0.38744466145833334, 0.38172526041666666, 0.38973958333333336, 0.3650618489583334, 0.382431640625, 0.3929622395833333, 0.3550553385416667, 0.4127864583333334, 0.3860416666666666, 0.355703125, 0.38600260416666665, 0.38702148437499995, 0.3895735677083333, 0.5193749999999999, 0.5799739583333333, 0.42284505208333334, 0.3665852864583333, 0.3497135416666667, 0.3780436197916667, 0.40083984375000004, 0.36480468749999995, 0.5630794270833333, 0.5853125, 0.388037109375, 0.38765950520833337, 0.40250976562499996, 0.3671126302083333, 0.4680924479166667, 0.385126953125, 0.36620768229166667, 0.37845377604166663, 0.4259505208333333, 0.3810904947916667, 0.3759440104166667, 0.39020507812500005, 0.41529947916666665, 0.384501953125, 0.377080078125, 0.39561848958333334, 0.3659537760416667, 0.38602213541666663, 0.36294921874999997, 0.4332552083333333, 0.4090950520833333, 0.40819986979166667, 0.5801009114583333, 0.5851334635416667, 0.39489908854166667, 0.3875065104166666, 0.381591796875, 0.5987076822916666, 0.3923795572916666, 0.363759765625, 0.38734049479166666, 0.4194791666666666, 0.36690755208333337, 0.3475716145833333, 0.3758463541666667]\n"
],
[
"series = pd.Series(data)\nseries.name = 'Data Density'\nprint(series.min())\nseries.head()",
"0.344488932292\n"
],
[
"bins = pd.cut(series,20)\nhistogram = bins.value_counts()\nprint(type(histogram))",
"<class 'pandas.core.series.Series'>\n"
],
[
"histogram.sort_index(inplace=True)\ntotal = sum(histogram)\nprint(total)\n",
"319\n"
],
[
"histogram.index",
"_____no_output_____"
],
[
"hist = [x/total for x in histogram]\nspan = series.max() - series.min()\nindex = np.linspace(series.min(),series.max(),len(hist))\nindex = map(lambda x: round(x,3),index)\nprint(index)\nhist = pd.Series(hist,index=index)",
"<map object at 0x112755be0>\n"
],
[
"plt.figure(\"Depth_Sensor_Performance\")\nhist.plot(kind='bar')\nplt.xlabel(\"Data Density\")\nplt.ylabel(\"Probability\")\nplt.title(\"Depth_Sensor_Performance: n=701,\")\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d003e6e33eceedc5e5cddd6fb58381fa9aead533 | 6,852 | ipynb | Jupyter Notebook | Samples/src/Arithmetic/Adder Example.ipynb | fafel/Quantum | 630c4ab7de5422c69b0a629740d231819fa88a49 | [
"MIT"
] | null | null | null | Samples/src/Arithmetic/Adder Example.ipynb | fafel/Quantum | 630c4ab7de5422c69b0a629740d231819fa88a49 | [
"MIT"
] | null | null | null | Samples/src/Arithmetic/Adder Example.ipynb | fafel/Quantum | 630c4ab7de5422c69b0a629740d231819fa88a49 | [
"MIT"
] | null | null | null | 25.662921 | 267 | 0.514594 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d003ef56f03dfdceaee65b794dacbe4d4ecf8265 | 48,437 | ipynb | Jupyter Notebook | Ch11_Optimization_Algorithms/11-9.ipynb | StevenJokess/d2l-en-read | 71b0f35971063b9fe5f21319b8072d61c9e5a298 | [
"MIT"
] | 1 | 2021-05-26T12:19:44.000Z | 2021-05-26T12:19:44.000Z | Ch11_Optimization_Algorithms/11-9.ipynb | StevenJokess/d2l-en-read | 71b0f35971063b9fe5f21319b8072d61c9e5a298 | [
"MIT"
] | null | null | null | Ch11_Optimization_Algorithms/11-9.ipynb | StevenJokess/d2l-en-read | 71b0f35971063b9fe5f21319b8072d61c9e5a298 | [
"MIT"
] | 1 | 2021-05-05T13:54:26.000Z | 2021-05-05T13:54:26.000Z | 393.796748 | 22,832 | 0.613993 | [
[
[
"%matplotlib inline\nfrom d2l import torch as d2l\nimport torch",
"_____no_output_____"
],
[
"def init_adadelta_states(feature_dim):\n s_w, s_b = torch.zeros((feature_dim, 1)), torch.zeros(1)\n delta_w, delta_b = torch.zeros((feature_dim, 1)), torch.zeros(1)\n return ((s_w, delta_w), (s_b, delta_b))",
"_____no_output_____"
],
[
"def adadelta(params, states, hyperparams):\n rho, eps = hyperparams['rho'], 1e-5\n for p, (s, delta) in zip(params, states):\n with torch.no_grad():\n # In-place updates via [:]\n s[:] = rho * s + (1 - rho) * torch.square(p.grad)\n g = (torch.sqrt(delta + eps) / torch.sqrt(s + eps)) * p.grad\n p[:] -= g\n delta[:] = rho * delta + (1 - rho) * g * g\n p.grad.data.zero_()",
"_____no_output_____"
],
[
"data_iter, feature_dim = d2l.get_data_ch11(batch_size=10)\nd2l.train_ch11(adadelta, init_adadelta_states(feature_dim),\n {'rho': 0.9}, data_iter, feature_dim);",
"loss: 0.243, 0.026 sec/epoch\n"
],
[
"trainer = torch.optim.Adadelta\nd2l.train_concise_ch11(trainer, {'rho': 0.9}, data_iter)",
"loss: 0.244, 0.023 sec/epoch\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d00405c26bbe9c7f2bef47f5af53566489e8e8a0 | 30,991 | ipynb | Jupyter Notebook | notebooks/.ipynb_checkpoints/Matrix-checkpoint.ipynb | raissabthibes/bmc | 840800fb94ea3bf188847d0771ca7197dfec68e3 | [
"MIT"
] | null | null | null | notebooks/.ipynb_checkpoints/Matrix-checkpoint.ipynb | raissabthibes/bmc | 840800fb94ea3bf188847d0771ca7197dfec68e3 | [
"MIT"
] | null | null | null | notebooks/.ipynb_checkpoints/Matrix-checkpoint.ipynb | raissabthibes/bmc | 840800fb94ea3bf188847d0771ca7197dfec68e3 | [
"MIT"
] | null | null | null | 26.53339 | 1,019 | 0.498984 | [
[
[
"# Matrix\n\n> Marcos Duarte \n> Laboratory of Biomechanics and Motor Control ([http://demotu.org/](http://demotu.org/)) \n> Federal University of ABC, Brazil",
"_____no_output_____"
],
[
"A matrix is a square or rectangular array of numbers or symbols (termed elements), arranged in rows and columns. For instance:\n\n$$ \n\\mathbf{A} = \n\\begin{bmatrix} \na_{1,1} & a_{1,2} & a_{1,3} \\\\\na_{2,1} & a_{2,2} & a_{2,3} \n\\end{bmatrix}\n$$\n\n$$ \n\\mathbf{A} = \n\\begin{bmatrix} \n1 & 2 & 3 \\\\\n4 & 5 & 6 \n\\end{bmatrix}\n$$\n\nThe matrix $\\mathbf{A}$ above has two rows and three columns, it is a 2x3 matrix.\n\nIn Numpy:",
"_____no_output_____"
]
],
[
[
"# Import the necessary libraries\nimport numpy as np\nfrom IPython.display import display\nnp.set_printoptions(precision=4) # number of digits of precision for floating point",
"_____no_output_____"
],
[
"A = np.array([[1, 2, 3], [4, 5, 6]])\nA",
"_____no_output_____"
]
],
[
[
"To get information about the number of elements and the structure of the matrix (in fact, a Numpy array), we can use:",
"_____no_output_____"
]
],
[
[
"print('A:\\n', A)\nprint('len(A) = ', len(A))\nprint('np.size(A) = ', np.size(A))\nprint('np.shape(A) = ', np.shape(A))\nprint('np.ndim(A) = ', np.ndim(A))",
"A:\n [[1 2 3]\n [4 5 6]]\nlen(A) = 2\nnp.size(A) = 6\nnp.shape(A) = (2, 3)\nnp.ndim(A) = 2\n"
]
],
[
[
"We could also have accessed this information with the correspondent methods:",
"_____no_output_____"
]
],
[
[
"print('A.size = ', A.size)\nprint('A.shape = ', A.shape)\nprint('A.ndim = ', A.ndim)",
"A.size = 6\nA.shape = (2, 3)\nA.ndim = 2\n"
]
],
[
[
"We used the array function in Numpy to represent a matrix. A [Numpy array is in fact different than a matrix](http://www.scipy.org/NumPy_for_Matlab_Users), if we want to use explicit matrices in Numpy, we have to use the function `mat`:",
"_____no_output_____"
]
],
[
[
"B = np.mat([[1, 2, 3], [4, 5, 6]])\nB",
"_____no_output_____"
]
],
[
[
"Both array and matrix types work in Numpy, but you should choose only one type and not mix them; the array is preferred because it is [the standard vector/matrix/tensor type of Numpy](http://www.scipy.org/NumPy_for_Matlab_Users). So, let's use the array type for the rest of this text.",
"_____no_output_____"
],
[
"## Addition and multiplication\n\nThe sum of two m-by-n matrices $\\mathbf{A}$ and $\\mathbf{B}$ is another m-by-n matrix:",
"_____no_output_____"
],
[
"$$ \n\\mathbf{A} = \n\\begin{bmatrix} \na_{1,1} & a_{1,2} & a_{1,3} \\\\\na_{2,1} & a_{2,2} & a_{2,3} \n\\end{bmatrix}\n\\;\\;\\; \\text{and} \\;\\;\\;\n\\mathbf{B} =\n\\begin{bmatrix} \nb_{1,1} & b_{1,2} & b_{1,3} \\\\\nb_{2,1} & b_{2,2} & b_{2,3} \n\\end{bmatrix}\n$$\n\n$$\n\\mathbf{A} + \\mathbf{B} = \n\\begin{bmatrix} \na_{1,1}+b_{1,1} & a_{1,2}+b_{1,2} & a_{1,3}+b_{1,3} \\\\\na_{2,1}+b_{2,1} & a_{2,2}+b_{2,2} & a_{2,3}+b_{2,3} \n\\end{bmatrix}\n$$\n\nIn Numpy:",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2, 3], [4, 5, 6]])\nB = np.array([[7, 8, 9], [10, 11, 12]])\nprint('A:\\n', A)\nprint('B:\\n', B)\nprint('A + B:\\n', A+B);",
"A:\n [[1 2 3]\n [4 5 6]]\nB:\n [[ 7 8 9]\n [10 11 12]]\nA + B:\n [[ 8 10 12]\n [14 16 18]]\n"
]
],
[
[
"The multiplication of the m-by-n matrix $\\mathbf{A}$ by the n-by-p matrix $\\mathbf{B}$ is a m-by-p matrix:\n\n$$ \n\\mathbf{A} = \n\\begin{bmatrix} \na_{1,1} & a_{1,2} \\\\\na_{2,1} & a_{2,2} \n\\end{bmatrix}\n\\;\\;\\; \\text{and} \\;\\;\\;\n\\mathbf{B} =\n\\begin{bmatrix} \nb_{1,1} & b_{1,2} & b_{1,3} \\\\\nb_{2,1} & b_{2,2} & b_{2,3} \n\\end{bmatrix}\n$$\n\n$$\n\\mathbf{A} \\mathbf{B} = \n\\begin{bmatrix} \na_{1,1}b_{1,1} + a_{1,2}b_{2,1} & a_{1,1}b_{1,2} + a_{1,2}b_{2,2} & a_{1,1}b_{1,3} + a_{1,2}b_{2,3} \\\\\na_{2,1}b_{1,1} + a_{2,2}b_{2,1} & a_{2,1}b_{1,2} + a_{2,2}b_{2,2} & a_{2,1}b_{1,3} + a_{2,2}b_{2,3}\n\\end{bmatrix}\n$$\n\nIn Numpy:",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2], [3, 4]])\nB = np.array([[5, 6, 7], [8, 9, 10]])\nprint('A:\\n', A)\nprint('B:\\n', B)\nprint('A x B:\\n', np.dot(A, B));",
"A:\n [[1 2]\n [3 4]]\nB:\n [[ 5 6 7]\n [ 8 9 10]]\nA x B:\n [[21 24 27]\n [47 54 61]]\n"
]
],
[
[
"Note that because the array type is not truly a matrix type, we used the dot product to calculate matrix multiplication. \nWe can use the matrix type to show the equivalent:",
"_____no_output_____"
]
],
[
[
"A = np.mat(A)\nB = np.mat(B)\nprint('A:\\n', A)\nprint('B:\\n', B)\nprint('A x B:\\n', A*B);",
"A:\n [[1 2]\n [3 4]]\nB:\n [[ 5 6 7]\n [ 8 9 10]]\nA x B:\n [[21 24 27]\n [47 54 61]]\n"
]
],
[
[
"Same result as before.\n\nThe order in multiplication matters, $\\mathbf{AB} \\neq \\mathbf{BA}$:",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2], [3, 4]])\nB = np.array([[5, 6], [7, 8]])\nprint('A:\\n', A)\nprint('B:\\n', B)\nprint('A x B:\\n', np.dot(A, B))\nprint('B x A:\\n', np.dot(B, A));",
"A:\n [[1 2]\n [3 4]]\nB:\n [[5 6]\n [7 8]]\nA x B:\n [[19 22]\n [43 50]]\nB x A:\n [[23 34]\n [31 46]]\n"
]
],
[
[
"The addition or multiplication of a scalar (a single number) to a matrix is performed over all the elements of the matrix:",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2], [3, 4]])\nc = 10\nprint('A:\\n', A)\nprint('c:\\n', c)\nprint('c + A:\\n', c+A)\nprint('cA:\\n', c*A);",
"A:\n [[1 2]\n [3 4]]\nc:\n 10\nc + A:\n [[11 12]\n [13 14]]\ncA:\n [[10 20]\n [30 40]]\n"
]
],
[
[
"## Transposition\n\nThe transpose of the matrix $\\mathbf{A}$ is the matrix $\\mathbf{A^T}$ turning all the rows of matrix $\\mathbf{A}$ into columns (or columns into rows):\n\n$$ \n\\mathbf{A} = \n\\begin{bmatrix} \na & b & c \\\\\nd & e & f \\end{bmatrix}\n\\;\\;\\;\\;\\;\\;\\iff\\;\\;\\;\\;\\;\\;\n\\mathbf{A^T} = \n\\begin{bmatrix} \na & d \\\\\nb & e \\\\\nc & f\n\\end{bmatrix} $$\n\nIn NumPy, the transpose operator can be used as a method or function:",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2], [3, 4]])\nprint('A:\\n', A)\nprint('A.T:\\n', A.T)\nprint('np.transpose(A):\\n', np.transpose(A));",
"A:\n [[1 2]\n [3 4]]\nA.T:\n [[1 3]\n [2 4]]\nnp.transpose(A):\n [[1 3]\n [2 4]]\n"
]
],
[
[
"## Determinant\n\nThe determinant is a number associated with a square matrix.\n\nThe determinant of the following matrix: \n\n$$ \\left[ \\begin{array}{ccc}\na & b & c \\\\\nd & e & f \\\\\ng & h & i \\end{array} \\right] $$\n\nis written as:\n\n$$ \\left| \\begin{array}{ccc}\na & b & c \\\\\nd & e & f \\\\\ng & h & i \\end{array} \\right| $$\n\nAnd has the value:\n\n$$ (aei + bfg + cdh) - (ceg + bdi + afh) $$\n\nOne way to manually calculate the determinant of a matrix is to use the [rule of Sarrus](http://en.wikipedia.org/wiki/Rule_of_Sarrus): we repeat the last columns (all columns but the first one) in the right side of the matrix and calculate the sum of the products of three diagonal north-west to south-east lines of matrix elements, minus the sum of the products of three diagonal south-west to north-east lines of elements as illustrated in the following figure: \n<br>\n<figure><img src='http://upload.wikimedia.org/wikipedia/commons/6/66/Sarrus_rule.svg' width=300 alt='Rule of Sarrus'/><center><figcaption><i>Figure. Rule of Sarrus: the sum of the products of the solid diagonals minus the sum of the products of the dashed diagonals (<a href=\"http://en.wikipedia.org/wiki/Rule_of_Sarrus\">image from Wikipedia</a>).</i></figcaption></center> </figure>\n\nIn Numpy, the determinant is computed with the `linalg.det` function:",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2], [3, 4]])\nprint('A:\\n', A);",
"A:\n [[1 2]\n [3 4]]\n"
],
[
"print('Determinant of A:\\n', np.linalg.det(A))",
"Determinant of A:\n -2.0\n"
]
],
[
[
"## Identity\n\nThe identity matrix $\\mathbf{I}$ is a matrix with ones in the main diagonal and zeros otherwise. The 3x3 identity matrix is: \n\n$$ \\mathbf{I} = \n\\begin{bmatrix} \n1 & 0 & 0 \\\\\n0 & 1 & 0 \\\\\n0 & 0 & 1 \\end{bmatrix} $$\n\nIn Numpy, instead of manually creating this matrix we can use the function `eye`:",
"_____no_output_____"
]
],
[
[
"np.eye(3) # identity 3x3 array",
"_____no_output_____"
]
],
[
[
"## Inverse\n\nThe inverse of the matrix $\\mathbf{A}$ is the matrix $\\mathbf{A^{-1}}$ such that the product between these two matrices is the identity matrix:\n\n$$ \\mathbf{A}\\cdot\\mathbf{A^{-1}} = \\mathbf{I} $$\n\nThe calculation of the inverse of a matrix is usually not simple (the inverse of the matrix $\\mathbf{A}$ is not $1/\\mathbf{A}$; there is no division operation between matrices). The Numpy function `linalg.inv` computes the inverse of a square matrix: \n\n numpy.linalg.inv(a)\n Compute the (multiplicative) inverse of a matrix.\n Given a square matrix a, return the matrix ainv satisfying dot(a, ainv) = dot(ainv, a) = eye(a.shape[0]).",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2], [3, 4]])\nprint('A:\\n', A)\nAinv = np.linalg.inv(A)\nprint('Inverse of A:\\n', Ainv);",
"A:\n [[1 2]\n [3 4]]\nInverse of A:\n [[-2. 1. ]\n [ 1.5 -0.5]]\n"
]
],
[
[
"### Pseudo-inverse\n\nFor a non-square matrix, its inverse is not defined. However, we can calculate what it's known as the pseudo-inverse. \nConsider a non-square matrix, $\\mathbf{A}$. To calculate its inverse, note that the following manipulation results in the identity matrix:\n\n$$ \\mathbf{A} \\mathbf{A}^T (\\mathbf{A}\\mathbf{A}^T)^{-1} = \\mathbf{I} $$\n\nThe $\\mathbf{A} \\mathbf{A}^T$ is a square matrix and is invertible (also [nonsingular](https://en.wikipedia.org/wiki/Invertible_matrix)) if $\\mathbf{A}$ is L.I. ([linearly independent rows/columns](https://en.wikipedia.org/wiki/Linear_independence)). \nThe matrix $\\mathbf{A}^T(\\mathbf{A}\\mathbf{A}^T)^{-1}$ is known as the [generalized inverse or Moore–Penrose pseudoinverse](https://en.wikipedia.org/wiki/Moore%E2%80%93Penrose_pseudoinverse) of the matrix $\\mathbf{A}$, a generalization of the inverse matrix.\n\nTo compute the Moore–Penrose pseudoinverse, we could calculate it by a naive approach in Python:\n```python\nfrom numpy.linalg import inv\nAinv = A.T @ inv(A @ A.T)\n```\nBut both Numpy and Scipy have functions to calculate the pseudoinverse, which might give greater numerical stability (but read [Inverses and pseudoinverses. Numerical issues, speed, symmetry](http://vene.ro/blog/inverses-pseudoinverses-numerical-issues-speed-symmetry.html)). Of note, [numpy.linalg.pinv](http://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.pinv.html) calculates the pseudoinverse of a matrix using its singular-value decomposition (SVD) and including all large singular values (using the [LAPACK (Linear Algebra Package)](https://en.wikipedia.org/wiki/LAPACK) routine gesdd), whereas [scipy.linalg.pinv](http://docs.scipy.org/doc/scipy/reference/generated/scipy.linalg.pinv.html#scipy.linalg.pinv) calculates a pseudoinverse of a matrix using a least-squares solver (using the LAPACK method gelsd) and [scipy.linalg.pinv2](http://docs.scipy.org/doc/scipy/reference/generated/scipy.linalg.pinv2.html) also uses SVD to find the pseudoinverse (also using the LAPACK routine gesdd). \n\nFor example:",
"_____no_output_____"
]
],
[
[
"from scipy.linalg import pinv2\n\nA = np.array([[1, 0, 0], [0, 1, 0]])\nApinv = pinv2(A)\nprint('Matrix A:\\n', A)\nprint('Pseudo-inverse of A:\\n', Apinv)\nprint('A x Apinv:\\n', A@Apinv)",
"Matrix A:\n [[1 0 0]\n [0 1 0]]\nPseudo-inverse of A:\n [[ 1. 0.]\n [ 0. 1.]\n [ 0. 0.]]\nA x Apinv:\n [[ 1. 0.]\n [ 0. 1.]]\n"
]
],
[
[
"## Orthogonality\n\nA square matrix is said to be orthogonal if:\n\n1. There is no linear combination of one of the lines or columns of the matrix that would lead to the other row or column. \n2. Its columns or rows form a basis of (independent) unit vectors (versors).\n\nAs consequence:\n\n1. Its determinant is equal to 1 or -1.\n2. Its inverse is equal to its transpose.\n\nHowever, keep in mind that not all matrices with determinant equals to one are orthogonal, for example, the matrix:\n\n$$ \\begin{bmatrix}\n3 & 2 \\\\\n4 & 3 \n\\end{bmatrix} $$\n\nHas determinant equals to one but it is not orthogonal (the columns or rows don't have norm equals to one).",
"_____no_output_____"
],
[
"## Linear equations\n\n> A linear equation is an algebraic equation in which each term is either a constant or the product of a constant and (the first power of) a single variable ([Wikipedia](http://en.wikipedia.org/wiki/Linear_equation)).\n\nWe are interested in solving a set of linear equations where two or more variables are unknown, for instance:\n\n$$ x + 2y = 4 $$\n\n$$ 3x + 4y = 10 $$\n\nLet's see how to employ the matrix formalism to solve these equations (even that we know the solution is `x=2` and `y=1`). \nLet's express this set of equations in matrix form:\n\n$$ \n\\begin{bmatrix} \n1 & 2 \\\\\n3 & 4 \\end{bmatrix}\n\\begin{bmatrix} \nx \\\\\ny \\end{bmatrix}\n= \\begin{bmatrix} \n4 \\\\\n10 \\end{bmatrix}\n$$\n\nAnd for the general case:\n\n$$ \\mathbf{Av} = \\mathbf{c} $$\n\nWhere $\\mathbf{A, v, c}$ are the matrices above and we want to find the values `x,y` for the matrix $\\mathbf{v}$. \nBecause there is no division of matrices, we can use the inverse of $\\mathbf{A}$ to solve for $\\mathbf{v}$:\n\n$$ \\mathbf{A}^{-1}\\mathbf{Av} = \\mathbf{A}^{-1}\\mathbf{c} \\implies $$\n\n$$ \\mathbf{v} = \\mathbf{A}^{-1}\\mathbf{c} $$\n\nAs we know how to compute the inverse of $\\mathbf{A}$, the solution is:",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2], [3, 4]])\nAinv = np.linalg.inv(A)\nc = np.array([4, 10])\nv = np.dot(Ainv, c)\nprint('v:\\n', v)",
"v:\n [ 2. 1.]\n"
]
],
[
[
"What we expected.\n\nHowever, the use of the inverse of a matrix to solve equations is computationally inefficient. \nInstead, we should use `linalg.solve` for a determined system (same number of equations and unknowns) or `linalg.lstsq` otherwise: \nFrom the help for `solve`: \n\n numpy.linalg.solve(a, b)[source]\n Solve a linear matrix equation, or system of linear scalar equations.\n Computes the “exact” solution, x, of the well-determined, i.e., full rank, linear matrix equation ax = b.",
"_____no_output_____"
]
],
[
[
"v = np.linalg.solve(A, c)\nprint('Using solve:')\nprint('v:\\n', v)",
"Using solve:\nv:\n [ 2. 1.]\n"
]
],
[
[
"And from the help for `lstsq`:\n\n numpy.linalg.lstsq(a, b, rcond=-1)[source]\n Return the least-squares solution to a linear matrix equation.\n Solves the equation a x = b by computing a vector x that minimizes the Euclidean 2-norm || b - a x ||^2. The equation may be under-, well-, or over- determined (i.e., the number of linearly independent rows of a can be less than, equal to, or greater than its number of linearly independent columns). If a is square and of full rank, then x (but for round-off error) is the “exact” solution of the equation.",
"_____no_output_____"
]
],
[
[
"v = np.linalg.lstsq(A, c)[0]\nprint('Using lstsq:')\nprint('v:\\n', v)",
"Using lstsq:\nv:\n [ 2. 1.]\n"
]
],
[
[
"Same solutions, of course.\n\nWhen a system of equations has a unique solution, the determinant of the **square** matrix associated to this system of equations is nonzero. \nWhen the determinant is zero there are either no solutions or many solutions to the system of equations.\n\nBut if we have an overdetermined system:\n\n$$ x + 2y = 4 $$\n\n$$ 3x + 4y = 10 $$\n\n$$ 5x + 6y = 15 $$\n\n(Note that the possible solution for this set of equations is not exact because the last equation should be equal to 16.)\n\nLet's try to solve it:",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2], [3, 4], [5, 6]])\nprint('A:\\n', A)\nc = np.array([4, 10, 15])\nprint('c:\\n', c);",
"A:\n [[1 2]\n [3 4]\n [5 6]]\nc:\n [ 4 10 15]\n"
]
],
[
[
"Because the matix $\\mathbf{A}$ is not squared, we can calculate its pseudo-inverse or use the function `linalg.lstsq`:",
"_____no_output_____"
]
],
[
[
"v = np.linalg.lstsq(A, c)[0]\nprint('Using lstsq:')\nprint('v:\\n', v)",
"Using lstsq:\nv:\n [ 1.3333 1.4167]\n"
]
],
[
[
"The functions `inv` and `solve` failed because the matrix $\\mathbf{A}$ was not square (overdetermined system). The function `lstsq` not only was able to handle an overdetermined system but was also able to find the best approximate solution.\n\nAnd if the the set of equations was undetermined, `lstsq` would also work. For instance, consider the system:\n\n$$ x + 2y + 2z = 10 $$\n\n$$ 3x + 4y + z = 13 $$\n\nAnd in matrix form:\n\n$$ \n\\begin{bmatrix} \n1 & 2 & 2 \\\\\n3 & 4 & 1 \\end{bmatrix}\n\\begin{bmatrix} \nx \\\\\ny \\\\\nz \\end{bmatrix}\n= \\begin{bmatrix} \n10 \\\\\n13 \\end{bmatrix}\n$$\n\nA possible solution would be `x=2,y=1,z=3`, but other values would also satisfy this set of equations.\n\nLet's try to solve using `lstsq`:",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, 2, 2], [3, 4, 1]])\nprint('A:\\n', A)\nc = np.array([10, 13])\nprint('c:\\n', c);",
"A:\n [[1 2 2]\n [3 4 1]]\nc:\n [10 13]\n"
],
[
"v = np.linalg.lstsq(A, c)[0]\nprint('Using lstsq:')\nprint('v:\\n', v);",
"Using lstsq:\nv:\n [ 0.8 2. 2.6]\n"
]
],
[
[
"This is an approximated solution and as explained in the help of `solve`, this solution, `v`, is the one that minimizes the Euclidean norm $|| \\mathbf{c - A v} ||^2$.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d004070b4f7fc66b6d7f11146a3c0cefbaa72435 | 162,625 | ipynb | Jupyter Notebook | embedding_word_clusters2.ipynb | mzkhan2000/KG-Embeddings | a56cc9df706817e05346fb9a2083b87d4bd27380 | [
"MIT"
] | null | null | null | embedding_word_clusters2.ipynb | mzkhan2000/KG-Embeddings | a56cc9df706817e05346fb9a2083b87d4bd27380 | [
"MIT"
] | null | null | null | embedding_word_clusters2.ipynb | mzkhan2000/KG-Embeddings | a56cc9df706817e05346fb9a2083b87d4bd27380 | [
"MIT"
] | null | null | null | 27.913663 | 25,572 | 0.317958 | [
[
[
"<a href=\"https://colab.research.google.com/github/mzkhan2000/KG-Embeddings/blob/main/embedding_word_clusters2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# Python program to generate embedding (word vectors) using Word2Vec\n\n# importing necessary modules for embedding\n!pip install --upgrade gensim",
"Requirement already satisfied: gensim in /usr/local/lib/python3.7/dist-packages (3.6.0)\nCollecting gensim\n Downloading gensim-4.1.0-cp37-cp37m-manylinux_2_12_x86_64.manylinux2010_x86_64.whl (24.0 MB)\n\u001b[K |████████████████████████████████| 24.0 MB 80 kB/s \n\u001b[?25hRequirement already satisfied: scipy>=0.18.1 in /usr/local/lib/python3.7/dist-packages (from gensim) (1.4.1)\nRequirement already satisfied: numpy>=1.17.0 in /usr/local/lib/python3.7/dist-packages (from gensim) (1.19.5)\nRequirement already satisfied: smart-open>=1.8.1 in /usr/local/lib/python3.7/dist-packages (from gensim) (5.1.0)\nInstalling collected packages: gensim\n Attempting uninstall: gensim\n Found existing installation: gensim 3.6.0\n Uninstalling gensim-3.6.0:\n Successfully uninstalled gensim-3.6.0\nSuccessfully installed gensim-4.1.0\n"
],
[
"!pip install rdflib\nimport rdflib",
"Collecting rdflib\n Downloading rdflib-6.0.0-py3-none-any.whl (376 kB)\n\u001b[?25l\r\u001b[K |▉ | 10 kB 16.9 MB/s eta 0:00:01\r\u001b[K |█▊ | 20 kB 20.0 MB/s eta 0:00:01\r\u001b[K |██▋ | 30 kB 23.5 MB/s eta 0:00:01\r\u001b[K |███▌ | 40 kB 25.6 MB/s eta 0:00:01\r\u001b[K |████▍ | 51 kB 9.4 MB/s eta 0:00:01\r\u001b[K |█████▏ | 61 kB 9.3 MB/s eta 0:00:01\r\u001b[K |██████ | 71 kB 8.4 MB/s eta 0:00:01\r\u001b[K |███████ | 81 kB 9.3 MB/s eta 0:00:01\r\u001b[K |███████▉ | 92 kB 9.4 MB/s eta 0:00:01\r\u001b[K |████████▊ | 102 kB 8.2 MB/s eta 0:00:01\r\u001b[K |█████████▋ | 112 kB 8.2 MB/s eta 0:00:01\r\u001b[K |██████████▍ | 122 kB 8.2 MB/s eta 0:00:01\r\u001b[K |███████████▎ | 133 kB 8.2 MB/s eta 0:00:01\r\u001b[K |████████████▏ | 143 kB 8.2 MB/s eta 0:00:01\r\u001b[K |█████████████ | 153 kB 8.2 MB/s eta 0:00:01\r\u001b[K |██████████████ | 163 kB 8.2 MB/s eta 0:00:01\r\u001b[K |██████████████▉ | 174 kB 8.2 MB/s eta 0:00:01\r\u001b[K |███████████████▋ | 184 kB 8.2 MB/s eta 0:00:01\r\u001b[K |████████████████▌ | 194 kB 8.2 MB/s eta 0:00:01\r\u001b[K |█████████████████▍ | 204 kB 8.2 MB/s eta 0:00:01\r\u001b[K |██████████████████▎ | 215 kB 8.2 MB/s eta 0:00:01\r\u001b[K |███████████████████▏ | 225 kB 8.2 MB/s eta 0:00:01\r\u001b[K |████████████████████ | 235 kB 8.2 MB/s eta 0:00:01\r\u001b[K |████████████████████▉ | 245 kB 8.2 MB/s eta 0:00:01\r\u001b[K |█████████████████████▊ | 256 kB 8.2 MB/s eta 0:00:01\r\u001b[K |██████████████████████▋ | 266 kB 8.2 MB/s eta 0:00:01\r\u001b[K |███████████████████████▌ | 276 kB 8.2 MB/s eta 0:00:01\r\u001b[K |████████████████████████▍ | 286 kB 8.2 MB/s eta 0:00:01\r\u001b[K |█████████████████████████▎ | 296 kB 8.2 MB/s eta 0:00:01\r\u001b[K |██████████████████████████ | 307 kB 8.2 MB/s eta 0:00:01\r\u001b[K |███████████████████████████ | 317 kB 8.2 MB/s eta 0:00:01\r\u001b[K |███████████████████████████▉ | 327 kB 8.2 MB/s eta 0:00:01\r\u001b[K |████████████████████████████▊ | 337 kB 8.2 MB/s eta 0:00:01\r\u001b[K |█████████████████████████████▋ | 348 kB 8.2 MB/s eta 0:00:01\r\u001b[K |██████████████████████████████▌ | 358 kB 8.2 MB/s eta 0:00:01\r\u001b[K |███████████████████████████████▎| 368 kB 8.2 MB/s eta 0:00:01\r\u001b[K |████████████████████████████████| 376 kB 8.2 MB/s \n\u001b[?25hRequirement already satisfied: setuptools in /usr/local/lib/python3.7/dist-packages (from rdflib) (57.4.0)\nRequirement already satisfied: pyparsing in /usr/local/lib/python3.7/dist-packages (from rdflib) (2.4.7)\nCollecting isodate\n Downloading isodate-0.6.0-py2.py3-none-any.whl (45 kB)\n\u001b[K |████████████████████████████████| 45 kB 2.9 MB/s \n\u001b[?25hRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from isodate->rdflib) (1.15.0)\nInstalling collected packages: isodate, rdflib\nSuccessfully installed isodate-0.6.0 rdflib-6.0.0\n"
],
[
"!pip uninstall numpy",
"Found existing installation: numpy 1.19.5\nUninstalling numpy-1.19.5:\n Would remove:\n /usr/bin/f2py\n /usr/local/bin/f2py\n /usr/local/bin/f2py3\n /usr/local/bin/f2py3.7\n /usr/local/lib/python3.7/dist-packages/numpy-1.19.5.dist-info/*\n /usr/local/lib/python3.7/dist-packages/numpy.libs/libgfortran-2e0d59d6.so.5.0.0\n /usr/local/lib/python3.7/dist-packages/numpy.libs/libopenblasp-r0-09e95953.3.13.so\n /usr/local/lib/python3.7/dist-packages/numpy.libs/libquadmath-2d0c479f.so.0.0.0\n /usr/local/lib/python3.7/dist-packages/numpy.libs/libz-eb09ad1d.so.1.2.3\n /usr/local/lib/python3.7/dist-packages/numpy/*\nProceed (y/n)? y\n Successfully uninstalled numpy-1.19.5\n"
],
[
"!pip install numpy",
"Collecting numpy\n Downloading numpy-1.21.2-cp37-cp37m-manylinux_2_12_x86_64.manylinux2010_x86_64.whl (15.7 MB)\n\u001b[K |████████████████████████████████| 15.7 MB 178 kB/s \n\u001b[?25hInstalling collected packages: numpy\n\u001b[31mERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.\ntensorflow 2.6.0 requires numpy~=1.19.2, but you have numpy 1.21.2 which is incompatible.\ndatascience 0.10.6 requires folium==0.2.1, but you have folium 0.8.3 which is incompatible.\nalbumentations 0.1.12 requires imgaug<0.2.7,>=0.2.5, but you have imgaug 0.2.9 which is incompatible.\u001b[0m\nSuccessfully installed numpy-1.21.2\n"
],
[
"# pip install numpy and then hit the RESTART RUNTIME\nimport gensim\nfrom gensim.models import Word2Vec\nfrom gensim.models import KeyedVectors\nfrom gensim.scripts.glove2word2vec import glove2word2vec\n\nimport collections\nfrom collections import Counter\n\nfrom rdflib import Graph, URIRef, Namespace",
"_____no_output_____"
],
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Mounted at /content/drive\n"
],
[
"# check out if google dride mount suceessful \n!ls \"/content/drive/My Drive/MonirResearchDatasets\"",
"all1k.ttl.txt\nFreebase-GoogleNews-vectors.bin\nfreebase-vectors-skipgram1000-en.bin\nGlove-6B\ngoogle_benchmark_data_embeddings_evaluation\ngoogle_benchmark_data_embeddings_evaluation_update\ngoogle-question-words.txt\nGravity_DBpedia.txt\nsurround-ga-records\n"
],
[
"# a funtion for ga-themes extraction from GA-rdf repository separate and return a list all the ga-themes - Monir\ndef gaThemesExtraction(ga_record):\n gaThemes = []\n with open(ga_record, 'rt') as f:\n data = f.readlines()\n for line in data:\n # check if line contains \"ga-themes\" sub-string\n if line.__contains__('ga-themes'):\n # split the line contains from \"ga-themes\" sub-string\n stringTemp = line.split(\"ga-themes/\",1)[1]\n # further split the line contains from \"ga-themes\" sub-string to delimiter\n stringTemp = stringTemp.split('>')[0]\n gaThemes.append(stringTemp)\n #print(dataLog)\n #print(gaThemes[:9])\n #print(len(gaThemes))\n return gaThemes",
"_____no_output_____"
],
[
"# a funtion imput a list of ga-themes and return a list of unique ga-themes and another list of duplicate gaThemes - \ndef make_unique_gaThemes(list_all_ga_themes):\n # find a a list of unique ga-themes\n unique_gaThemes = []\n unique_gaThemes = list(dict.fromkeys(gaThemes))\n #print(len(unique_gaThemes))\n\n # a list of duplicate gaThemes\n duplicate_gaThemes = []\n duplicate_gaThemes = [item for item, count in collections.Counter(gaThemes).items() if count > 1]\n #print(len(duplicate_gaThemes))\n\n return unique_gaThemes, duplicate_gaThemes",
"_____no_output_____"
],
[
"## KG-Embeddings\nfilename = '/content/drive/My Drive/MonirResearchDatasets/Freebase-GoogleNews-vectors.bin'\nmodel = KeyedVectors.load_word2vec_format(filename, binary=True)",
"_____no_output_____"
],
[
"def embedding_word_clusters(model, list_of_ga_themes, cluster_size):\n keys = list_of_ga_themes\n embedding_model = model\n n = cluster_size\n new_classifier = []\n embedding_clusters = []\n classifier_clusters = []\n for word in keys:\n embeddings = []\n words = []\n # check if a word is fully \"OOV\" (out of vocabulary) for pre-trained embedding model\n if word in embedding_model.key_to_index:\n # create a new list of classifier\n new_classifier.append(word)\n # find most similar top n words from the pre-trained embedding model\n for similar_word, _ in embedding_model.most_similar(word, topn=n):\n words.append(similar_word)\n embeddings.append(embedding_model[similar_word])\n embedding_clusters.append(embeddings)\n classifier_clusters.append(words)\n\n return embedding_clusters, classifier_clusters, new_classifier",
"_____no_output_____"
],
[
"# to get all the ga-themes from all1K file \nga_record_datapath = \"/content/drive/My Drive/MonirResearchDatasets/surround-ga-records/all1k.ttl.txt\"\ngaThemes = gaThemesExtraction(ga_record_datapath)\nprint(gaThemes[:10])\nprint(len(gaThemes))",
"['palaeontology', 'geophysics', 'palaeontology', 'geophysics', 'palaeontology', 'stratigraphy', 'palaeontology', 'earthquakes', 'palaeontology', 'palaeontology']\n1204\n"
],
[
"# to get all unique ga-themes\nunique_gaThemes, duplicate_gaThemes = make_unique_gaThemes(gaThemes)\nprint(unique_gaThemes[:100])\n#print(duplicate_gaThemes[:100])\nprint(len(unique_gaThemes))",
"['palaeontology', 'geophysics', 'stratigraphy', 'earthquakes', 'geomagnetism', 'seismics', 'volcanology', 'groundwater', 'gravity', 'marine', 'magnetics', 'gamma-spectroscopy', 'geology', 'mineral-deposits', 'sedimentary-basins', 'metallogenesis', 'mineragraphy', 'petrography', 'mineral-exploration', 'hydrogeology', 'petroleum-exploration', 'landslides', 'geochemistry', 'petrology', 'marine-survey', 'economic-geology', 'petroleum-geology', 'cartography', 'petroleum-reserves', 'remote-sensing', 'structural-geology', 'continental-margins', 'geochronology', 'bathymetry', 'palaeogeography', 'geoscience-databases', 'data-standards', 'gis', 'landforms', 'regolith', 'metamorphism', 'image-processing', 'satellite-imagery', 'palaeomagnetism', 'geoscience-education', 'tsunamis', 'seismology', 'information-management', 'marine-jurisdiction', 'coasts', 'geomorphology', 'resource-management', 'mineralogy', 'sequence-stratigraphy', 'seismic-sections', 'mapping', 'administrative-boundaries', 'boundaries', 'dataset', 'national-dataset', 'topography', 'water-resources', 'hydrocarbons', 'aem', 'dating', 'educational-product', 'environmental', 'carbon-dioxide', 'co2-capture', 'geological-sequestration', 'geological-storage-of-co2', 'risk-assessment', 'carbonate', 'geodesy', 'geodynamics', 'model', 'resource-assessment', 'pmd*crc', 'tourism', 'geothermal', 'marine-zone', 'commodities', 'culture', '3d-model', 'geohazards', 'geoscience', 'numerical-modelling', 'airborne-electromagnetics', 'land-cover', 'magneto-tellurics', 'soils', 'atmosphere', 'abiotic-surrogates', 'seismic-velocity', 'seabed', 'minerals', 'energy-infrastructure', 'risk-analysis', 'transport', 'infrastructure']\n124\n"
],
[
"embedding_clusters, classifier_clusters, new_classifier = embedding_word_clusters(model, unique_gaThemes[:10], 10)",
"_____no_output_____"
],
[
"print(classifier_clusters)",
"[['paleontology', 'paleontological', 'palaeontologists', 'paleoanthropology', 'palaeontologist', 'palaeontological', 'archeology', 'archaeo', 'paleobiology', 'paleontologist'], ['geophysical', 'geophysical_surveys', 'geophysical_survey', 'geological_mapping', 'geochemistry', 'geologic_mapping', 'geochemical', 'Geophysics', 'airborne_geophysics', 'detailed_geological_mapping'], ['volcanics', 'volcanic_stratigraphy', 'lithologies', 'shear_zone', 'stratigraphic', 'gold_mineralization', 'intrusive_rocks', 'intrusives', 'auriferous', 'meta_sediments'], ['quakes', 'temblors', 'earthquake', 'temblor', 'quake', '#.#_magnitude_earthquake', 'tsunamis', 'quakes_measuring', 'seismic_activity', 'aftershocks'], ['Michael_Studinger', 'Cosmic_Background_Explorer_COBE', 'polarimetric', 'Anisotropy_Probe', 'glaciological', 'exoplanetary', 'radar_interferometry', 'meteorological_oceanographic', 'atmospheric_oceanic', 'SeaWiFS'], ['4D_seismic', '3D_seismic_surveys', 'Induced_Polarisation', 'magnetic_geophysical_surveys', '2D_seismic_surveys', '2D_seismic_data', 'dimensional_seismic', '2D_seismic_survey', 'Triassic_reservoirs', 'resistivity_surveying'], ['vulcanology', 'seismology', 'vulcanologist', 'Syamsu_Rizal', 'volcano_observatory', 'volcanology_institute', 'Volcanology', 'Agus_Budianto', 'volcanologist', 'Umar_Rosadi'], ['aquifer', 'aquifers', 'Groundwater', 'shallow_aquifer', 'underground_aquifer', 'underground_aquifers', 'groundwater_aquifers', 'groundwater_aquifer', 'shallow_aquifers', 'Aquifer'], ['gravitational', 'gravitational_pull', 'Hejlik_MRAP_request', 'angular_momentum', 'gravitation', 'rotational_axis', 'gravitational_acceleration', 'gravitational_pulls', 'centrifugal_force', 'gravitational_attraction'], ['Marine', 'maritime', 'marine_mammal', 'fisheries', 'WolfCamera.com', 'coral_reef', 'vessel_groundings', 'marine_mammals', 'marine_ecology', 'seafloor_habitats']]\n"
],
[
"print(new_classifier)",
"['palaeontology', 'geophysics', 'stratigraphy', 'earthquakes', 'geomagnetism', 'seismics', 'volcanology', 'groundwater', 'gravity', 'marine']\n"
],
[
"print(classifier_clusters[:2])",
"[['paleontology', 'paleontological', 'palaeontologists', 'paleoanthropology', 'palaeontologist', 'palaeontological', 'archeology', 'archaeo', 'paleobiology', 'paleontologist'], ['geophysical', 'geophysical_surveys', 'geophysical_survey', 'geological_mapping', 'geochemistry', 'geologic_mapping', 'geochemical', 'Geophysics', 'airborne_geophysics', 'detailed_geological_mapping']]\n"
],
[
"print(new_classifier[:2])",
"['palaeontology', 'geophysics']\n"
],
[
"from rdflib import Graph\n\ng = Graph()\ng.parse(\"/content/drive/My Drive/MonirResearchDatasets/surround-ga-records/ga-records.ttl\", format='turtle')\n\nprint(len(g))",
"843765\n"
],
[
"n_record = Namespace(\"http://example.com/record/\")\n# <http://example.com/record/105030>\nn_GA = Namespace(\"http://example.org/def/ga-themes/\")\nn_hasClassifier = Namespace(\"http://data.surroundaustralia.com/def/agr#\")\n\nhasClassifier = \"hasClassifier\"",
"_____no_output_____"
],
[
"#record = []\nfor obj in new_classifier[:1]: # for obj in new_classifier:\n\n results = g.query(\n \"\"\"\n PREFIX classifier: <http://data.surroundaustralia.com/def/agr#>\n PREFIX ga-themes: <http://example.org/def/ga-themes/>\n \n SELECT ?s WHERE { ?s classifier:hasClassifier ga-themes:\"\"\" + obj + \"\"\" }\n \"\"\")\n\n record = []\n pos = new_classifier.index(obj)\n\n for row in results:\n # print(f\"{row.s}\")\n record.append(row.s)\n\n # adding classifier from classifier cluster to each of the list of records\n for classifier_obj in classifier_clusters[pos]:\n for record_data in record:\n g.add((record_data, n_hasClassifier.hasClassifier, n_GA[classifier_obj]))\n\n \n\n\n",
"_____no_output_____"
],
[
" \n # adding classifier from classifier cluster to the list of records\n for q in record:\n g.add((record[q], n_hasClassifier.hasClassifier, n_GA[classifier_clusters[1]]))",
"_____no_output_____"
],
[
"print(record)",
"[rdflib.term.URIRef('http://example.com/record/9730'), rdflib.term.URIRef('http://example.com/record/9833'), rdflib.term.URIRef('http://example.com/record/9621'), rdflib.term.URIRef('http://example.com/record/10317'), rdflib.term.URIRef('http://example.com/record/10086'), rdflib.term.URIRef('http://example.com/record/9543'), rdflib.term.URIRef('http://example.com/record/22975'), rdflib.term.URIRef('http://example.com/record/10206'), rdflib.term.URIRef('http://example.com/record/10290'), rdflib.term.URIRef('http://example.com/record/9495'), rdflib.term.URIRef('http://example.com/record/9947'), rdflib.term.URIRef('http://example.com/record/10010'), rdflib.term.URIRef('http://example.com/record/14938'), rdflib.term.URIRef('http://example.com/record/10263'), rdflib.term.URIRef('http://example.com/record/9437'), rdflib.term.URIRef('http://example.com/record/10548'), rdflib.term.URIRef('http://example.com/record/41'), rdflib.term.URIRef('http://example.com/record/68164'), rdflib.term.URIRef('http://example.com/record/14272'), rdflib.term.URIRef('http://example.com/record/96'), rdflib.term.URIRef('http://example.com/record/34'), rdflib.term.URIRef('http://example.com/record/10273'), rdflib.term.URIRef('http://example.com/record/12327'), rdflib.term.URIRef('http://example.com/record/9817'), rdflib.term.URIRef('http://example.com/record/9906'), rdflib.term.URIRef('http://example.com/record/9846'), rdflib.term.URIRef('http://example.com/record/9442'), rdflib.term.URIRef('http://example.com/record/9693'), rdflib.term.URIRef('http://example.com/record/9782'), rdflib.term.URIRef('http://example.com/record/41716'), rdflib.term.URIRef('http://example.com/record/10307'), rdflib.term.URIRef('http://example.com/record/9872'), rdflib.term.URIRef('http://example.com/record/11249'), rdflib.term.URIRef('http://example.com/record/9631'), rdflib.term.URIRef('http://example.com/record/9720'), rdflib.term.URIRef('http://example.com/record/9640'), rdflib.term.URIRef('http://example.com/record/9943'), rdflib.term.URIRef('http://example.com/record/10045'), rdflib.term.URIRef('http://example.com/record/9866'), rdflib.term.URIRef('http://example.com/record/8979'), rdflib.term.URIRef('http://example.com/record/71568'), rdflib.term.URIRef('http://example.com/record/14242'), rdflib.term.URIRef('http://example.com/record/92'), rdflib.term.URIRef('http://example.com/record/14384'), rdflib.term.URIRef('http://example.com/record/9359'), rdflib.term.URIRef('http://example.com/record/9786'), rdflib.term.URIRef('http://example.com/record/9446'), rdflib.term.URIRef('http://example.com/record/10183'), rdflib.term.URIRef('http://example.com/record/9724'), rdflib.term.URIRef('http://example.com/record/9664'), rdflib.term.URIRef('http://example.com/record/9936'), rdflib.term.URIRef('http://example.com/record/9472'), rdflib.term.URIRef('http://example.com/record/10226'), rdflib.term.URIRef('http://example.com/record/9563'), rdflib.term.URIRef('http://example.com/record/9423'), rdflib.term.URIRef('http://example.com/record/10055'), rdflib.term.URIRef('http://example.com/record/9601'), rdflib.term.URIRef('http://example.com/record/10115'), rdflib.term.URIRef('http://example.com/record/9813'), rdflib.term.URIRef('http://example.com/record/9481'), rdflib.term.URIRef('http://example.com/record/9710'), rdflib.term.URIRef('http://example.com/record/9641'), rdflib.term.URIRef('http://example.com/record/10044'), rdflib.term.URIRef('http://example.com/record/9432'), rdflib.term.URIRef('http://example.com/record/9572'), rdflib.term.URIRef('http://example.com/record/9463'), rdflib.term.URIRef('http://example.com/record/9523'), rdflib.term.URIRef('http://example.com/record/148'), rdflib.term.URIRef('http://example.com/record/9624'), rdflib.term.URIRef('http://example.com/record/9764'), rdflib.term.URIRef('http://example.com/record/9517'), rdflib.term.URIRef('http://example.com/record/9457'), rdflib.term.URIRef('http://example.com/record/9348'), rdflib.term.URIRef('http://example.com/record/15'), rdflib.term.URIRef('http://example.com/record/11452'), rdflib.term.URIRef('http://example.com/record/70'), rdflib.term.URIRef('http://example.com/record/14267'), rdflib.term.URIRef('http://example.com/record/12839'), rdflib.term.URIRef('http://example.com/record/9218'), rdflib.term.URIRef('http://example.com/record/9634'), rdflib.term.URIRef('http://example.com/record/9422'), rdflib.term.URIRef('http://example.com/record/12373'), rdflib.term.URIRef('http://example.com/record/9903'), rdflib.term.URIRef('http://example.com/record/9600'), rdflib.term.URIRef('http://example.com/record/10114'), rdflib.term.URIRef('http://example.com/record/9952'), rdflib.term.URIRef('http://example.com/record/9711'), rdflib.term.URIRef('http://example.com/record/9480'), rdflib.term.URIRef('http://example.com/record/9671'), rdflib.term.URIRef('http://example.com/record/9832'), rdflib.term.URIRef('http://example.com/record/9542'), rdflib.term.URIRef('http://example.com/record/10316'), rdflib.term.URIRef('http://example.com/record/10196'), rdflib.term.URIRef('http://example.com/record/9857'), rdflib.term.URIRef('http://example.com/record/12145'), rdflib.term.URIRef('http://example.com/record/9494'), rdflib.term.URIRef('http://example.com/record/9946'), rdflib.term.URIRef('http://example.com/record/14939'), rdflib.term.URIRef('http://example.com/record/14273'), rdflib.term.URIRef('http://example.com/record/72990'), rdflib.term.URIRef('http://example.com/record/15208'), rdflib.term.URIRef('http://example.com/record/64'), rdflib.term.URIRef('http://example.com/record/9566'), rdflib.term.URIRef('http://example.com/record/9537'), rdflib.term.URIRef('http://example.com/record/9816'), rdflib.term.URIRef('http://example.com/record/10141'), rdflib.term.URIRef('http://example.com/record/168'), rdflib.term.URIRef('http://example.com/record/9907'), rdflib.term.URIRef('http://example.com/record/9503'), rdflib.term.URIRef('http://example.com/record/9692'), rdflib.term.URIRef('http://example.com/record/9783'), rdflib.term.URIRef('http://example.com/record/9880'), rdflib.term.URIRef('http://example.com/record/9630'), rdflib.term.URIRef('http://example.com/record/9721'), rdflib.term.URIRef('http://example.com/record/9100'), rdflib.term.URIRef('http://example.com/record/10467'), rdflib.term.URIRef('http://example.com/record/9233'), rdflib.term.URIRef('http://example.com/record/9322'), rdflib.term.URIRef('http://example.com/record/10402'), rdflib.term.URIRef('http://example.com/record/12447'), rdflib.term.URIRef('http://example.com/record/9196'), rdflib.term.URIRef('http://example.com/record/10682'), rdflib.term.URIRef('http://example.com/record/9518'), rdflib.term.URIRef('http://example.com/record/116'), rdflib.term.URIRef('http://example.com/record/8984'), rdflib.term.URIRef('http://example.com/record/9928'), rdflib.term.URIRef('http://example.com/record/240'), rdflib.term.URIRef('http://example.com/record/10269'), rdflib.term.URIRef('http://example.com/record/180'), rdflib.term.URIRef('http://example.com/record/173'), rdflib.term.URIRef('http://example.com/record/132'), rdflib.term.URIRef('http://example.com/record/61127'), rdflib.term.URIRef('http://example.com/record/250'), rdflib.term.URIRef('http://example.com/record/11004'), rdflib.term.URIRef('http://example.com/record/10228'), rdflib.term.URIRef('http://example.com/record/13257'), rdflib.term.URIRef('http://example.com/record/9419'), rdflib.term.URIRef('http://example.com/record/10873'), rdflib.term.URIRef('http://example.com/record/9124'), rdflib.term.URIRef('http://example.com/record/14268'), rdflib.term.URIRef('http://example.com/record/10847'), rdflib.term.URIRef('http://example.com/record/9050'), rdflib.term.URIRef('http://example.com/record/15110'), rdflib.term.URIRef('http://example.com/record/10686'), rdflib.term.URIRef('http://example.com/record/9104'), rdflib.term.URIRef('http://example.com/record/10523'), rdflib.term.URIRef('http://example.com/record/9377'), rdflib.term.URIRef('http://example.com/record/14967'), rdflib.term.URIRef('http://example.com/record/244'), rdflib.term.URIRef('http://example.com/record/14557'), rdflib.term.URIRef('http://example.com/record/68802'), rdflib.term.URIRef('http://example.com/record/9858'), rdflib.term.URIRef('http://example.com/record/13212'), rdflib.term.URIRef('http://example.com/record/8980'), rdflib.term.URIRef('http://example.com/record/8990'), rdflib.term.URIRef('http://example.com/record/14611'), rdflib.term.URIRef('http://example.com/record/9908'), rdflib.term.URIRef('http://example.com/record/14984'), rdflib.term.URIRef('http://example.com/record/136'), rdflib.term.URIRef('http://example.com/record/9819'), rdflib.term.URIRef('http://example.com/record/9538'), rdflib.term.URIRef('http://example.com/record/13948'), rdflib.term.URIRef('http://example.com/record/9285'), rdflib.term.URIRef('http://example.com/record/9054'), rdflib.term.URIRef('http://example.com/record/9093'), rdflib.term.URIRef('http://example.com/record/9242'), rdflib.term.URIRef('http://example.com/record/9213'), rdflib.term.URIRef('http://example.com/record/10665'), rdflib.term.URIRef('http://example.com/record/9060'), rdflib.term.URIRef('http://example.com/record/12972'), rdflib.term.URIRef('http://example.com/record/23168'), rdflib.term.URIRef('http://example.com/record/37468'), rdflib.term.URIRef('http://example.com/record/9342'), rdflib.term.URIRef('http://example.com/record/13569'), rdflib.term.URIRef('http://example.com/record/10700'), rdflib.term.URIRef('http://example.com/record/9154'), rdflib.term.URIRef('http://example.com/record/9294'), rdflib.term.URIRef('http://example.com/record/15216'), rdflib.term.URIRef('http://example.com/record/10573'), rdflib.term.URIRef('http://example.com/record/9438'), rdflib.term.URIRef('http://example.com/record/13145'), rdflib.term.URIRef('http://example.com/record/185'), rdflib.term.URIRef('http://example.com/record/11380'), rdflib.term.URIRef('http://example.com/record/245'), rdflib.term.URIRef('http://example.com/record/176'), rdflib.term.URIRef('http://example.com/record/14843'), rdflib.term.URIRef('http://example.com/record/10198'), rdflib.term.URIRef('http://example.com/record/142'), rdflib.term.URIRef('http://example.com/record/8981'), rdflib.term.URIRef('http://example.com/record/13203'), rdflib.term.URIRef('http://example.com/record/152'), rdflib.term.URIRef('http://example.com/record/10308'), rdflib.term.URIRef('http://example.com/record/230'), rdflib.term.URIRef('http://example.com/record/11064'), rdflib.term.URIRef('http://example.com/record/166'), rdflib.term.URIRef('http://example.com/record/9818'), rdflib.term.URIRef('http://example.com/record/13326'), rdflib.term.URIRef('http://example.com/record/9539'), rdflib.term.URIRef('http://example.com/record/14867'), rdflib.term.URIRef('http://example.com/record/14927'), rdflib.term.URIRef('http://example.com/record/9479'), rdflib.term.URIRef('http://example.com/record/14735'), rdflib.term.URIRef('http://example.com/record/10472'), rdflib.term.URIRef('http://example.com/record/9337'), rdflib.term.URIRef('http://example.com/record/9055'), rdflib.term.URIRef('http://example.com/record/15075'), rdflib.term.URIRef('http://example.com/record/9144'), rdflib.term.URIRef('http://example.com/record/9303'), rdflib.term.URIRef('http://example.com/record/9101'), rdflib.term.URIRef('http://example.com/record/9290'), rdflib.term.URIRef('http://example.com/record/73734'), rdflib.term.URIRef('http://example.com/record/9010'), rdflib.term.URIRef('http://example.com/record/15061'), rdflib.term.URIRef('http://example.com/record/9372'), rdflib.term.URIRef('http://example.com/record/15183'), rdflib.term.URIRef('http://example.com/record/10621'), rdflib.term.URIRef('http://example.com/record/9086'), rdflib.term.URIRef('http://example.com/record/9346'), rdflib.term.URIRef('http://example.com/record/224'), rdflib.term.URIRef('http://example.com/record/9408'), rdflib.term.URIRef('http://example.com/record/11130'), rdflib.term.URIRef('http://example.com/record/9519'), rdflib.term.URIRef('http://example.com/record/9929'), rdflib.term.URIRef('http://example.com/record/10268'), rdflib.term.URIRef('http://example.com/record/172'), rdflib.term.URIRef('http://example.com/record/133'), rdflib.term.URIRef('http://example.com/record/191'), rdflib.term.URIRef('http://example.com/record/14620'), rdflib.term.URIRef('http://example.com/record/9879'), rdflib.term.URIRef('http://example.com/record/156'), rdflib.term.URIRef('http://example.com/record/107'), rdflib.term.URIRef('http://example.com/record/14917'), rdflib.term.URIRef('http://example.com/record/14269'), rdflib.term.URIRef('http://example.com/record/9222'), rdflib.term.URIRef('http://example.com/record/9140'), rdflib.term.URIRef('http://example.com/record/10817'), rdflib.term.URIRef('http://example.com/record/9250'), rdflib.term.URIRef('http://example.com/record/9201'), rdflib.term.URIRef('http://example.com/record/9341'), rdflib.term.URIRef('http://example.com/record/9023'), rdflib.term.URIRef('http://example.com/record/9072'), rdflib.term.URIRef('http://example.com/record/10570'), rdflib.term.URIRef('http://example.com/record/13117'), rdflib.term.URIRef('http://example.com/record/186'), rdflib.term.URIRef('http://example.com/record/11084'), rdflib.term.URIRef('http://example.com/record/141'), rdflib.term.URIRef('http://example.com/record/10139'), rdflib.term.URIRef('http://example.com/record/61366'), rdflib.term.URIRef('http://example.com/record/10178'), rdflib.term.URIRef('http://example.com/record/37224'), rdflib.term.URIRef('http://example.com/record/11113'), rdflib.term.URIRef('http://example.com/record/134'), rdflib.term.URIRef('http://example.com/record/12298'), rdflib.term.URIRef('http://example.com/record/9658'), rdflib.term.URIRef('http://example.com/record/14986'), rdflib.term.URIRef('http://example.com/record/15136'), rdflib.term.URIRef('http://example.com/record/9116'), rdflib.term.URIRef('http://example.com/record/9056'), rdflib.term.URIRef('http://example.com/record/10531'), rdflib.term.URIRef('http://example.com/record/9365'), rdflib.term.URIRef('http://example.com/record/9300'), rdflib.term.URIRef('http://example.com/record/9371'), rdflib.term.URIRef('http://example.com/record/10525'), rdflib.term.URIRef('http://example.com/record/10465'), rdflib.term.URIRef('http://example.com/record/10762'), rdflib.term.URIRef('http://example.com/record/10733'), rdflib.term.URIRef('http://example.com/record/12875'), rdflib.term.URIRef('http://example.com/record/9027'), rdflib.term.URIRef('http://example.com/record/14844'), rdflib.term.URIRef('http://example.com/record/8975'), rdflib.term.URIRef('http://example.com/record/14955'), rdflib.term.URIRef('http://example.com/record/13360'), rdflib.term.URIRef('http://example.com/record/242'), rdflib.term.URIRef('http://example.com/record/252'), rdflib.term.URIRef('http://example.com/record/10148'), rdflib.term.URIRef('http://example.com/record/9998'), rdflib.term.URIRef('http://example.com/record/9728'), rdflib.term.URIRef('http://example.com/record/9668'), rdflib.term.URIRef('http://example.com/record/8996'), rdflib.term.URIRef('http://example.com/record/9639'), rdflib.term.URIRef('http://example.com/record/12925'), rdflib.term.URIRef('http://example.com/record/9304'), rdflib.term.URIRef('http://example.com/record/59'), rdflib.term.URIRef('http://example.com/record/9361'), rdflib.term.URIRef('http://example.com/record/12461'), rdflib.term.URIRef('http://example.com/record/15210'), rdflib.term.URIRef('http://example.com/record/9012'), rdflib.term.URIRef('http://example.com/record/12712'), rdflib.term.URIRef('http://example.com/record/10617'), rdflib.term.URIRef('http://example.com/record/48'), rdflib.term.URIRef('http://example.com/record/10510'), rdflib.term.URIRef('http://example.com/record/9344'), rdflib.term.URIRef('http://example.com/record/10401'), rdflib.term.URIRef('http://example.com/record/15057'), rdflib.term.URIRef('http://example.com/record/61011'), rdflib.term.URIRef('http://example.com/record/9628'), rdflib.term.URIRef('http://example.com/record/9768'), rdflib.term.URIRef('http://example.com/record/70225'), rdflib.term.URIRef('http://example.com/record/12128'), rdflib.term.URIRef('http://example.com/record/14845'), rdflib.term.URIRef('http://example.com/record/10019'), rdflib.term.URIRef('http://example.com/record/170'), rdflib.term.URIRef('http://example.com/record/253'), rdflib.term.URIRef('http://example.com/record/11116'), rdflib.term.URIRef('http://example.com/record/160'), rdflib.term.URIRef('http://example.com/record/11225'), rdflib.term.URIRef('http://example.com/record/9729'), rdflib.term.URIRef('http://example.com/record/8997'), rdflib.term.URIRef('http://example.com/record/9669'), rdflib.term.URIRef('http://example.com/record/154'), rdflib.term.URIRef('http://example.com/record/9305'), rdflib.term.URIRef('http://example.com/record/9094'), rdflib.term.URIRef('http://example.com/record/10747'), rdflib.term.URIRef('http://example.com/record/9113'), rdflib.term.URIRef('http://example.com/record/9142'), rdflib.term.URIRef('http://example.com/record/10736'), rdflib.term.URIRef('http://example.com/record/9133'), rdflib.term.URIRef('http://example.com/record/13598'), rdflib.term.URIRef('http://example.com/record/15214'), rdflib.term.URIRef('http://example.com/record/10613'), rdflib.term.URIRef('http://example.com/record/9618'), rdflib.term.URIRef('http://example.com/record/174'), rdflib.term.URIRef('http://example.com/record/9709'), rdflib.term.URIRef('http://example.com/record/9498'), rdflib.term.URIRef('http://example.com/record/68801'), rdflib.term.URIRef('http://example.com/record/247'), rdflib.term.URIRef('http://example.com/record/14875'), rdflib.term.URIRef('http://example.com/record/64962'), rdflib.term.URIRef('http://example.com/record/140'), rdflib.term.URIRef('http://example.com/record/14841'), rdflib.term.URIRef('http://example.com/record/11066'), rdflib.term.URIRef('http://example.com/record/10128'), rdflib.term.URIRef('http://example.com/record/70371'), rdflib.term.URIRef('http://example.com/record/10068'), rdflib.term.URIRef('http://example.com/record/150'), rdflib.term.URIRef('http://example.com/record/10179'), rdflib.term.URIRef('http://example.com/record/8993'), rdflib.term.URIRef('http://example.com/record/101'), rdflib.term.URIRef('http://example.com/record/14737'), rdflib.term.URIRef('http://example.com/record/9659'), rdflib.term.URIRef('http://example.com/record/9719'), rdflib.term.URIRef('http://example.com/record/9608'), rdflib.term.URIRef('http://example.com/record/39'), rdflib.term.URIRef('http://example.com/record/10530'), rdflib.term.URIRef('http://example.com/record/10874'), rdflib.term.URIRef('http://example.com/record/11639'), rdflib.term.URIRef('http://example.com/record/10265'), rdflib.term.URIRef('http://example.com/record/9571'), rdflib.term.URIRef('http://example.com/record/12393'), rdflib.term.URIRef('http://example.com/record/9613'), rdflib.term.URIRef('http://example.com/record/9642'), rdflib.term.URIRef('http://example.com/record/10080'), rdflib.term.URIRef('http://example.com/record/229'), rdflib.term.URIRef('http://example.com/record/9405'), rdflib.term.URIRef('http://example.com/record/9685'), rdflib.term.URIRef('http://example.com/record/10340'), rdflib.term.URIRef('http://example.com/record/13069'), rdflib.term.URIRef('http://example.com/record/10200'), rdflib.term.URIRef('http://example.com/record/9676'), rdflib.term.URIRef('http://example.com/record/9736'), rdflib.term.URIRef('http://example.com/record/20786'), rdflib.term.URIRef('http://example.com/record/9078'), rdflib.term.URIRef('http://example.com/record/10758'), rdflib.term.URIRef('http://example.com/record/63'), rdflib.term.URIRef('http://example.com/record/9068'), rdflib.term.URIRef('http://example.com/record/9934'), rdflib.term.URIRef('http://example.com/record/8998'), rdflib.term.URIRef('http://example.com/record/9666'), rdflib.term.URIRef('http://example.com/record/9996'), rdflib.term.URIRef('http://example.com/record/9887'), rdflib.term.URIRef('http://example.com/record/239'), rdflib.term.URIRef('http://example.com/record/9951'), rdflib.term.URIRef('http://example.com/record/9712'), rdflib.term.URIRef('http://example.com/record/12230'), rdflib.term.URIRef('http://example.com/record/9421'), rdflib.term.URIRef('http://example.com/record/9530'), rdflib.term.URIRef('http://example.com/record/10195'), rdflib.term.URIRef('http://example.com/record/11168'), rdflib.term.URIRef('http://example.com/record/9541'), rdflib.term.URIRef('http://example.com/record/10315'), rdflib.term.URIRef('http://example.com/record/9893'), rdflib.term.URIRef('http://example.com/record/10077'), rdflib.term.URIRef('http://example.com/record/9623'), rdflib.term.URIRef('http://example.com/record/69795'), rdflib.term.URIRef('http://example.com/record/188'), rdflib.term.URIRef('http://example.com/record/9464'), rdflib.term.URIRef('http://example.com/record/9945'), rdflib.term.URIRef('http://example.com/record/10103'), rdflib.term.URIRef('http://example.com/record/84'), rdflib.term.URIRef('http://example.com/record/26'), rdflib.term.URIRef('http://example.com/record/14270'), rdflib.term.URIRef('http://example.com/record/13663'), rdflib.term.URIRef('http://example.com/record/12'), rdflib.term.URIRef('http://example.com/record/9118'), rdflib.term.URIRef('http://example.com/record/9607'), rdflib.term.URIRef('http://example.com/record/9596'), rdflib.term.URIRef('http://example.com/record/9904'), rdflib.term.URIRef('http://example.com/record/9474'), rdflib.term.URIRef('http://example.com/record/9425'), rdflib.term.URIRef('http://example.com/record/69554'), rdflib.term.URIRef('http://example.com/record/9633'), rdflib.term.URIRef('http://example.com/record/9691'), rdflib.term.URIRef('http://example.com/record/9680'), rdflib.term.URIRef('http://example.com/record/10205'), rdflib.term.URIRef('http://example.com/record/9540'), rdflib.term.URIRef('http://example.com/record/9892'), rdflib.term.URIRef('http://example.com/record/9400'), rdflib.term.URIRef('http://example.com/record/10076'), rdflib.term.URIRef('http://example.com/record/9434'), rdflib.term.URIRef('http://example.com/record/9574'), rdflib.term.URIRef('http://example.com/record/13149'), rdflib.term.URIRef('http://example.com/record/12334'), rdflib.term.URIRef('http://example.com/record/10013'), rdflib.term.URIRef('http://example.com/record/10382'), rdflib.term.URIRef('http://example.com/record/12007'), rdflib.term.URIRef('http://example.com/record/10102'), rdflib.term.URIRef('http://example.com/record/9855'), rdflib.term.URIRef('http://example.com/record/9018'), rdflib.term.URIRef('http://example.com/record/37510'), rdflib.term.URIRef('http://example.com/record/9049'), rdflib.term.URIRef('http://example.com/record/63994'), rdflib.term.URIRef('http://example.com/record/13440'), rdflib.term.URIRef('http://example.com/record/14271'), rdflib.term.URIRef('http://example.com/record/11685'), rdflib.term.URIRef('http://example.com/record/42'), rdflib.term.URIRef('http://example.com/record/14383'), rdflib.term.URIRef('http://example.com/record/13646'), rdflib.term.URIRef('http://example.com/record/9597'), rdflib.term.URIRef('http://example.com/record/9657'), rdflib.term.URIRef('http://example.com/record/10330'), rdflib.term.URIRef('http://example.com/record/9424'), rdflib.term.URIRef('http://example.com/record/10270'), rdflib.term.URIRef('http://example.com/record/10177'), rdflib.term.URIRef('http://example.com/record/9663'), rdflib.term.URIRef('http://example.com/record/9632'), rdflib.term.URIRef('http://example.com/record/41956'), rdflib.term.URIRef('http://example.com/record/9882'), rdflib.term.URIRef('http://example.com/record/9781'), rdflib.term.URIRef('http://example.com/record/9441'), rdflib.term.URIRef('http://example.com/record/9570'), rdflib.term.URIRef('http://example.com/record/9612'), rdflib.term.URIRef('http://example.com/record/228'), rdflib.term.URIRef('http://example.com/record/10310'), rdflib.term.URIRef('http://example.com/record/9515'), rdflib.term.URIRef('http://example.com/record/9684'), rdflib.term.URIRef('http://example.com/record/65665'), rdflib.term.URIRef('http://example.com/record/9677'), rdflib.term.URIRef('http://example.com/record/9834'), rdflib.term.URIRef('http://example.com/record/9737'), rdflib.term.URIRef('http://example.com/record/9865'), rdflib.term.URIRef('http://example.com/record/15148'), rdflib.term.URIRef('http://example.com/record/10588'), rdflib.term.URIRef('http://example.com/record/70617'), rdflib.term.URIRef('http://example.com/record/23352'), rdflib.term.URIRef('http://example.com/record/9129'), rdflib.term.URIRef('http://example.com/record/11780'), rdflib.term.URIRef('http://example.com/record/9875'), rdflib.term.URIRef('http://example.com/record/9935'), rdflib.term.URIRef('http://example.com/record/9824'), rdflib.term.URIRef('http://example.com/record/9667'), rdflib.term.URIRef('http://example.com/record/10180'), rdflib.term.URIRef('http://example.com/record/12314'), rdflib.term.URIRef('http://example.com/record/9997'), rdflib.term.URIRef('http://example.com/record/9785'), rdflib.term.URIRef('http://example.com/record/9950'), rdflib.term.URIRef('http://example.com/record/9713'), rdflib.term.URIRef('http://example.com/record/9482'), rdflib.term.URIRef('http://example.com/record/9593'), rdflib.term.URIRef('http://example.com/record/9742'), rdflib.term.URIRef('http://example.com/record/10334'), rdflib.term.URIRef('http://example.com/record/10225')]\n"
],
[
"print(new_classifier)\n",
"['palaeontology', 'geophysics', 'stratigraphy', 'earthquakes', 'geomagnetism', 'seismics', 'volcanology', 'groundwater', 'gravity', 'marine']\n"
],
[
"print(new_classifier.index('palaeontology'))",
"0\n"
],
[
"print(classifier_clusters[0])",
"['paleontology', 'paleontological', 'palaeontologists', 'paleoanthropology', 'palaeontologist', 'palaeontological', 'archeology', 'archaeo', 'paleobiology', 'paleontologist']\n"
],
[
"print(len(record))",
"1232\n"
],
[
"print(len(record))",
"_____no_output_____"
],
[
"print(len(classifier_clusters))",
"10\n"
],
[
" a = [[1, 3, 4], [2, 4, 4], [3, 4, 5]]\n ",
"_____no_output_____"
],
[
"for recordlist in record:\n print(recordlist)\n for number in recordlist:\n print(number)",
"\u001b[1;30;43mStreaming output truncated to the last 5000 lines.\u001b[0m\ne\nc\no\nr\nd\n/\n1\n9\n1\nhttp://example.com/record/14620\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n6\n2\n0\nhttp://example.com/record/9879\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n8\n7\n9\nhttp://example.com/record/156\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n5\n6\nhttp://example.com/record/107\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n7\nhttp://example.com/record/14917\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n9\n1\n7\nhttp://example.com/record/14269\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n2\n6\n9\nhttp://example.com/record/9222\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n2\n2\n2\nhttp://example.com/record/9140\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n1\n4\n0\nhttp://example.com/record/10817\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n8\n1\n7\nhttp://example.com/record/9250\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n2\n5\n0\nhttp://example.com/record/9201\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n2\n0\n1\nhttp://example.com/record/9341\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n3\n4\n1\nhttp://example.com/record/9023\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n0\n2\n3\nhttp://example.com/record/9072\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n0\n7\n2\nhttp://example.com/record/10570\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n5\n7\n0\nhttp://example.com/record/13117\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n3\n1\n1\n7\nhttp://example.com/record/186\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n8\n6\nhttp://example.com/record/11084\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n1\n0\n8\n4\nhttp://example.com/record/141\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n1\nhttp://example.com/record/10139\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n1\n3\n9\nhttp://example.com/record/61366\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n6\n1\n3\n6\n6\nhttp://example.com/record/10178\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n1\n7\n8\nhttp://example.com/record/37224\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n3\n7\n2\n2\n4\nhttp://example.com/record/11113\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n1\n1\n1\n3\nhttp://example.com/record/134\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n3\n4\nhttp://example.com/record/12298\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n2\n2\n9\n8\nhttp://example.com/record/9658\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n5\n8\nhttp://example.com/record/14986\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n9\n8\n6\nhttp://example.com/record/15136\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n5\n1\n3\n6\nhttp://example.com/record/9116\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n1\n1\n6\nhttp://example.com/record/9056\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n0\n5\n6\nhttp://example.com/record/10531\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n5\n3\n1\nhttp://example.com/record/9365\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n3\n6\n5\nhttp://example.com/record/9300\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n3\n0\n0\nhttp://example.com/record/9371\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n3\n7\n1\nhttp://example.com/record/10525\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n5\n2\n5\nhttp://example.com/record/10465\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n4\n6\n5\nhttp://example.com/record/10762\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n7\n6\n2\nhttp://example.com/record/10733\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n7\n3\n3\nhttp://example.com/record/12875\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n2\n8\n7\n5\nhttp://example.com/record/9027\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n0\n2\n7\nhttp://example.com/record/14844\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n8\n4\n4\nhttp://example.com/record/8975\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n8\n9\n7\n5\nhttp://example.com/record/14955\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n9\n5\n5\nhttp://example.com/record/13360\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n3\n3\n6\n0\nhttp://example.com/record/242\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n2\n4\n2\nhttp://example.com/record/252\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n2\n5\n2\nhttp://example.com/record/10148\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n1\n4\n8\nhttp://example.com/record/9998\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n9\n9\n8\nhttp://example.com/record/9728\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n7\n2\n8\nhttp://example.com/record/9668\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n6\n8\nhttp://example.com/record/8996\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n8\n9\n9\n6\nhttp://example.com/record/9639\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n3\n9\nhttp://example.com/record/12925\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n2\n9\n2\n5\nhttp://example.com/record/9304\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n3\n0\n4\nhttp://example.com/record/59\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n5\n9\nhttp://example.com/record/9361\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n3\n6\n1\nhttp://example.com/record/12461\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n2\n4\n6\n1\nhttp://example.com/record/15210\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n5\n2\n1\n0\nhttp://example.com/record/9012\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n0\n1\n2\nhttp://example.com/record/12712\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n2\n7\n1\n2\nhttp://example.com/record/10617\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n6\n1\n7\nhttp://example.com/record/48\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n4\n8\nhttp://example.com/record/10510\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n5\n1\n0\nhttp://example.com/record/9344\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n3\n4\n4\nhttp://example.com/record/10401\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n4\n0\n1\nhttp://example.com/record/15057\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n5\n0\n5\n7\nhttp://example.com/record/61011\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n6\n1\n0\n1\n1\nhttp://example.com/record/9628\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n2\n8\nhttp://example.com/record/9768\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n7\n6\n8\nhttp://example.com/record/70225\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n7\n0\n2\n2\n5\nhttp://example.com/record/12128\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n2\n1\n2\n8\nhttp://example.com/record/14845\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n8\n4\n5\nhttp://example.com/record/10019\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n0\n1\n9\nhttp://example.com/record/170\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n7\n0\nhttp://example.com/record/253\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n2\n5\n3\nhttp://example.com/record/11116\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n1\n1\n1\n6\nhttp://example.com/record/160\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n6\n0\nhttp://example.com/record/11225\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n1\n2\n2\n5\nhttp://example.com/record/9729\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n7\n2\n9\nhttp://example.com/record/8997\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n8\n9\n9\n7\nhttp://example.com/record/9669\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n6\n9\nhttp://example.com/record/154\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n5\n4\nhttp://example.com/record/9305\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n3\n0\n5\nhttp://example.com/record/9094\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n0\n9\n4\nhttp://example.com/record/10747\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n7\n4\n7\nhttp://example.com/record/9113\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n1\n1\n3\nhttp://example.com/record/9142\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n1\n4\n2\nhttp://example.com/record/10736\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n7\n3\n6\nhttp://example.com/record/9133\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n1\n3\n3\nhttp://example.com/record/13598\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n3\n5\n9\n8\nhttp://example.com/record/15214\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n5\n2\n1\n4\nhttp://example.com/record/10613\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n6\n1\n3\nhttp://example.com/record/9618\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n1\n8\nhttp://example.com/record/174\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n7\n4\nhttp://example.com/record/9709\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n7\n0\n9\nhttp://example.com/record/9498\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n4\n9\n8\nhttp://example.com/record/68801\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n6\n8\n8\n0\n1\nhttp://example.com/record/247\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n2\n4\n7\nhttp://example.com/record/14875\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n8\n7\n5\nhttp://example.com/record/64962\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n6\n4\n9\n6\n2\nhttp://example.com/record/140\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n0\nhttp://example.com/record/14841\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n8\n4\n1\nhttp://example.com/record/11066\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n1\n0\n6\n6\nhttp://example.com/record/10128\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n1\n2\n8\nhttp://example.com/record/70371\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n7\n0\n3\n7\n1\nhttp://example.com/record/10068\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n0\n6\n8\nhttp://example.com/record/150\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n5\n0\nhttp://example.com/record/10179\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n1\n7\n9\nhttp://example.com/record/8993\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n8\n9\n9\n3\nhttp://example.com/record/101\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n1\nhttp://example.com/record/14737\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n4\n7\n3\n7\nhttp://example.com/record/9659\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n5\n9\nhttp://example.com/record/9719\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n7\n1\n9\nhttp://example.com/record/9608\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n0\n8\nhttp://example.com/record/39\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n3\n9\nhttp://example.com/record/10530\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n5\n3\n0\nhttp://example.com/record/10874\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n8\n7\n4\nhttp://example.com/record/11639\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n1\n6\n3\n9\nhttp://example.com/record/10265\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n2\n6\n5\nhttp://example.com/record/9571\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n5\n7\n1\nhttp://example.com/record/12393\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n2\n3\n9\n3\nhttp://example.com/record/9613\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n1\n3\nhttp://example.com/record/9642\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n4\n2\nhttp://example.com/record/10080\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n0\n8\n0\nhttp://example.com/record/229\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n2\n2\n9\nhttp://example.com/record/9405\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n4\n0\n5\nhttp://example.com/record/9685\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n8\n5\nhttp://example.com/record/10340\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n3\n4\n0\nhttp://example.com/record/13069\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n3\n0\n6\n9\nhttp://example.com/record/10200\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n2\n0\n0\nhttp://example.com/record/9676\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n7\n6\nhttp://example.com/record/9736\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n7\n3\n6\nhttp://example.com/record/20786\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n2\n0\n7\n8\n6\nhttp://example.com/record/9078\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n0\n7\n8\nhttp://example.com/record/10758\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n7\n5\n8\nhttp://example.com/record/63\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n6\n3\nhttp://example.com/record/9068\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n0\n6\n8\nhttp://example.com/record/9934\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n9\n3\n4\nhttp://example.com/record/8998\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n8\n9\n9\n8\nhttp://example.com/record/9666\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n6\n6\nhttp://example.com/record/9996\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n9\n9\n6\nhttp://example.com/record/9887\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n8\n8\n7\nhttp://example.com/record/239\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n2\n3\n9\nhttp://example.com/record/9951\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n9\n5\n1\nhttp://example.com/record/9712\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n7\n1\n2\nhttp://example.com/record/12230\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n2\n2\n3\n0\nhttp://example.com/record/9421\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n4\n2\n1\nhttp://example.com/record/9530\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n5\n3\n0\nhttp://example.com/record/10195\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n1\n9\n5\nhttp://example.com/record/11168\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n1\n1\n6\n8\nhttp://example.com/record/9541\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n5\n4\n1\nhttp://example.com/record/10315\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n3\n1\n5\nhttp://example.com/record/9893\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n8\n9\n3\nhttp://example.com/record/10077\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n0\n0\n7\n7\nhttp://example.com/record/9623\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n6\n2\n3\nhttp://example.com/record/69795\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n6\n9\n7\n9\n5\nhttp://example.com/record/188\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n1\n8\n8\nhttp://example.com/record/9464\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n.\nc\no\nm\n/\nr\ne\nc\no\nr\nd\n/\n9\n4\n6\n4\nhttp://example.com/record/9945\nh\nt\nt\np\n:\n/\n/\ne\nx\na\nm\np\nl\ne\n."
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0040e8578098f6f8b611f8d9e09479a0aab9a68 | 103,332 | ipynb | Jupyter Notebook | DAY-12/DAY-12.ipynb | BhuvaneshHingal/LetsUpgrade-AI-ML | 63f7114d680b2738c9c40983996adafe55c0edd2 | [
"MIT"
] | 1 | 2020-09-11T18:11:54.000Z | 2020-09-11T18:11:54.000Z | DAY-12/DAY-12.ipynb | BhuvaneshHingal/LetsUpgrade-AI-ML | 63f7114d680b2738c9c40983996adafe55c0edd2 | [
"MIT"
] | null | null | null | DAY-12/DAY-12.ipynb | BhuvaneshHingal/LetsUpgrade-AI-ML | 63f7114d680b2738c9c40983996adafe55c0edd2 | [
"MIT"
] | 1 | 2020-07-22T19:47:15.000Z | 2020-07-22T19:47:15.000Z | 41.632554 | 7,496 | 0.459277 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\ndataset1=pd.read_csv('general_data.csv')",
"_____no_output_____"
],
[
"dataset1.head()",
"_____no_output_____"
],
[
"dataset1.columns",
"_____no_output_____"
],
[
"dataset1",
"_____no_output_____"
],
[
"dataset1.isnull()",
"_____no_output_____"
],
[
"dataset1.duplicated()",
"_____no_output_____"
],
[
"dataset1.drop_duplicates()",
"_____no_output_____"
],
[
"dataset3=dataset1[['Age','DistanceFromHome','Education','MonthlyIncome', 'NumCompaniesWorked', 'PercentSalaryHike','TotalWorkingYears', 'TrainingTimesLastYear', 'YearsAtCompany','YearsSinceLastPromotion', 'YearsWithCurrManager']].describe()\ndataset3",
"_____no_output_____"
],
[
"dataset3=dataset1[['Age','DistanceFromHome','Education','MonthlyIncome', 'NumCompaniesWorked', 'PercentSalaryHike','TotalWorkingYears', 'TrainingTimesLastYear', 'YearsAtCompany','YearsSinceLastPromotion', 'YearsWithCurrManager']].median()\ndataset3",
"_____no_output_____"
],
[
"dataset3=dataset1[['Age','DistanceFromHome','Education','MonthlyIncome', 'NumCompaniesWorked', 'PercentSalaryHike','TotalWorkingYears', 'TrainingTimesLastYear', 'YearsAtCompany','YearsSinceLastPromotion', 'YearsWithCurrManager']].mode()\ndataset3",
"_____no_output_____"
],
[
"dataset3=dataset1[['Age','DistanceFromHome','Education','MonthlyIncome', 'NumCompaniesWorked', 'PercentSalaryHike','TotalWorkingYears', 'TrainingTimesLastYear', 'YearsAtCompany','YearsSinceLastPromotion', 'YearsWithCurrManager']].var()\ndataset3",
"_____no_output_____"
],
[
"dataset3=dataset1[['Age','DistanceFromHome','Education','MonthlyIncome', 'NumCompaniesWorked', 'PercentSalaryHike','TotalWorkingYears', 'TrainingTimesLastYear', 'YearsAtCompany','YearsSinceLastPromotion', 'YearsWithCurrManager']].skew()\ndataset3",
"_____no_output_____"
],
[
"dataset3=dataset1[['Age','DistanceFromHome','Education','MonthlyIncome', 'NumCompaniesWorked', 'PercentSalaryHike','TotalWorkingYears', 'TrainingTimesLastYear', 'YearsAtCompany','YearsSinceLastPromotion', 'YearsWithCurrManager']].kurt()\ndataset3",
"_____no_output_____"
]
],
[
[
"# Inference from the analysis:\n All the above variables show positive skewness; while Age & Mean_distance_from_home are leptokurtic and all other variables are platykurtic.\n\n The Mean_Monthly_Income’s IQR is at 54K suggesting company wide attrition across all income bands\n\n Mean age forms a near normal distribution with 13 years of IQR\n",
"_____no_output_____"
],
[
"# Outliers:\nThere’s no regression found while plotting Age, MonthlyIncome, TotalWorkingYears, YearsAtCompany, etc., on a scatter plot\n",
"_____no_output_____"
]
],
[
[
"box_plot=dataset1.Age\nplt.boxplot(box_plot)",
"_____no_output_____"
]
],
[
[
"### Age is normally distributed without any outliers",
"_____no_output_____"
]
],
[
[
"box_plot=dataset1.MonthlyIncome\nplt.boxplot(box_plot)",
"_____no_output_____"
]
],
[
[
"### Monthly Income is Right skewed with several outliers",
"_____no_output_____"
]
],
[
[
"box_plot=dataset1.YearsAtCompany\nplt.boxplot(box_plot)",
"_____no_output_____"
]
],
[
[
"### Years at company is also Right Skewed with several outliers observed.",
"_____no_output_____"
],
[
"# Attrition Vs Distance from Home",
"_____no_output_____"
]
],
[
[
"from scipy.stats import mannwhitneyu",
"_____no_output_____"
],
[
"from scipy.stats import mannwhitneyu\na1=dataset.DistanceFromHome_Yes\na2=dataset.DistanceFromHome_No\nstat, p=mannwhitneyu(a1,a2)\nprint(stat, p)\n3132625.5 0.0",
"_____no_output_____"
]
],
[
[
"As the P value of 0.0 is < 0.05, the H0 is rejected and Ha is accepted.\n\nH0: There is no significant differences in the Distance From Home between attrition (Y) and attirition (N)\n\nHa: There is significant differences in the Distance From Home between attrition (Y) and attirition (N)",
"_____no_output_____"
],
[
"## Attrition Vs Income",
"_____no_output_____"
]
],
[
[
"a1=dataset.MonthlyIncome_Yes\na2=dataset.MonthlyIncome_No\nstat, p=mannwhitneyu(a1,a2)\nprint(stat, p)\n3085416.0 0.0",
"_____no_output_____"
]
],
[
[
"As the P value is again 0.0, which is < than 0.05, the H0 is rejected and ha is accepted.\n\nH0: There is no significant differences in the income between attrition (Y) and attirition (N)\n\nHa: There is significant differences in the income between attrition (Y) and attirition (N)",
"_____no_output_____"
],
[
"## Attrition Vs Total Working Years",
"_____no_output_____"
]
],
[
[
"a1=dataset.TotalWorkingYears_Yes\na2=dataset.TotalWorkingYears_No\nstat, p=mannwhitneyu(a1,a2)\nprint(stat, p)\n2760982.0 0.0",
"_____no_output_____"
]
],
[
[
"As the P value is again 0.0, which is < than 0.05, the H0 is rejected and ha is accepted.\n\nH0: There is no significant differences in the Total Working Years between attrition (Y) and attirition (N)\n\nHa: There is significant differences in the Total Working Years between attrition (Y) and attirition (N)",
"_____no_output_____"
],
[
"## Attrition Vs Years at company",
"_____no_output_____"
]
],
[
[
"a1=dataset.YearsAtCompany_Yes\na2=dataset.YearsAtCompany_No\nstat, p=mannwhitneyu(a1,a2)\nprint(stat, p)\n2882047.5 0.0",
"_____no_output_____"
]
],
[
[
"As the P value is again 0.0, which is < than 0.05, the H0 is rejected and ha is accepted.\n\nH0: There is no significant differences in the Years At Company between attrition (Y) and attirition (N)\n \nHa: There is significant differences in the Years At Company between attrition (Y) and attirition (N)",
"_____no_output_____"
],
[
"## Attrition Vs YearsWithCurrentManager",
"_____no_output_____"
]
],
[
[
"a1=dataset.YearsWithCurrManager_Yes\na2=dataset.YearsWithCurrManager_No\nstat, p=mannwhitneyu(a1,a2)\nprint(stat, p)\n3674749.5 0.0",
"_____no_output_____"
]
],
[
[
"As the P value is again 0.0, which is < than 0.05, the H0 is rejected and ha is accepted.\n\nH0: There is no significant differences in the Years With Current Manager between attrition (Y) and attirition (N)\n\nHa: There is significant differences in the Years With Current Manager between attrition (Y) and attirition (N)",
"_____no_output_____"
],
[
"# Statistical Tests (Separate T Test)",
"_____no_output_____"
],
[
"## Attrition Vs Distance From Home\nfrom scipy.stats import ttest_ind",
"_____no_output_____"
]
],
[
[
"z1=dataset.DistanceFromHome_Yes\nz2=dataset.DistanceFromHome_No\nstat, p=ttest_ind(z2,z1)\nprint(stat, p)\n44.45445917636664 0.0",
"_____no_output_____"
]
],
[
[
"As the P value is again 0.0, which is < than 0.05, the H0 is rejected and ha is accepted.\n\nH0: There is no significant differences in the Distance From Home between attrition (Y) and attirition (N)\n\nHa: There is significant differences in the Distance From Home between attrition (Y) and attirition (N)",
"_____no_output_____"
],
[
"## Attrition Vs Income",
"_____no_output_____"
]
],
[
[
"z1=dataset.MonthlyIncome_Yes\nz2=dataset.MonthlyIncome_No\nstat, p=ttest_ind(z2, z1)\nprint(stat, p)\n52.09279408504947 0.0",
"_____no_output_____"
]
],
[
[
"As the P value is again 0.0, which is < than 0.05, the H0 is rejected and ha is accepted.\n\nH0: There is no significant differences in the Monthly Income between attrition (Y) and attirition (N)\n \nHa: There is significant differences in the Monthly Income between attrition (Y) and attirition (N)",
"_____no_output_____"
],
[
"## Attrition Vs Yeats At Company",
"_____no_output_____"
]
],
[
[
"z1=dataset.YearsAtCompany_Yes\nz2=dataset.YearsAtCompany_No\nstat, p=ttest_ind(z2, z1)\nprint(stat, p)\n51.45296941515692 0.0",
"_____no_output_____"
]
],
[
[
"As the P value is again 0.0, which is < than 0.05, the H0 is rejected and ha is accepted.\n\nH0: There is no significant differences in the Years At Company between attrition (Y) and attirition (N)\n\nHa: There is significant differences in the Years At Company between attrition (Y) and attirition (N)",
"_____no_output_____"
],
[
"## Attrition Vs Years With Current Manager",
"_____no_output_____"
]
],
[
[
"z1=dataset.YearsWithCurrManager_Yes\nz2=dataset.YearsWithCurrManager_No\nstat, p=ttest_ind(z2, z1)\nprint(stat, p)\n53.02424349024521 0.0",
"_____no_output_____"
]
],
[
[
"As the P value is again 0.0, which is < than 0.05, the H0 is rejected and ha is accepted.\n\nH0: There is no significant differences in the Years With Current Manager between attrition (Y) and attirition (N)\n\nHa: There is significant differences in the Years With Current Manager between attrition (Y) and attirition (N)",
"_____no_output_____"
],
[
"# Unsupervised Learning - Correlation Analysis\nIn order to find the interdependency of the variables DistanceFromHome, MonthlyIncome, TotalWorkingYears, YearsAtCompany, YearsWithCurrManager from that of Attrition, we executed the Correlation Analysis as follows.",
"_____no_output_____"
],
[
"stats, p=pearsonr(dataset.Attrition, dataset.DistanceFromHome)\n\nprint(stats, p)\n\n-0.009730141010179438 0.5182860428049617\n\nstats, p=pearsonr(dataset.Attrition, dataset.MonthlyIncome)\n\nprint(stats, p)\n\n-0.031176281698114025 0.0384274849060192\n\nstats, p=pearsonr(dataset.Attrition, dataset.TotalWorkingYears)\n\nprint(stats, p)\n\n-0.17011136355964646 5.4731597518148054e-30\n\nstats, p=pearsonr(dataset.Attrition, dataset.YearsAtCompany)\n\nprint(stats, p)\n\n-0.13439221398997386 3.163883122493571e-19\n\nstats, p=pearsonr(dataset.Attrition, dataset.YearsWithCurrManager)\n\nprint(stats, p)\n\n-0.15619931590162422 1.7339322652951965e-25",
"_____no_output_____"
],
[
"# The inference of the above analysis are as follows:",
"_____no_output_____"
],
[
"Attrition & DistanceFromHome:\n\nAs r = -0.009, there’s low negative correlation between Attrition and DistanceFromHome\n\nAs the P value of 0.518 is > 0.05, we are accepting H0 and hence there’s no significant correlation between Attrition & \n\nDistanceFromHome\n\nAttrition & MonthlyIncome:\n\nAs r = -0.031, there’s low negative correlation between Attrition and MonthlyIncome\n\nAs the P value of 0.038 is < 0.05, we are accepting Ha and hence there’s significant correlation between Attrition & \n\nMonthlyIncome\n\nAttrition & TotalWorkingYears:\n\nAs r = -0.17, there’s low negative correlation between Attrition and TotalWorkingYears\n\nAs the P value is < 0.05, we are accepting Ha and hence there’s significant correlation between Attrition & TotalWorkingYears\n\nAttrition & YearsAtCompany:\n\nAs r = -0.1343, there’s low negative correlation between Attrition and YearsAtCompany\n\nAs the P value is < 0.05, we are accepting Ha and hence there’s significant correlation between Attrition & YearsAtCompany\n\nAttrition & YearsWithCurrManager:\n\nAs r = -0.1561, there’s low negative correlation between Attrition and YearsWithCurrManager\n\nAs the P value is < 0.05, we are accepting Ha and hence there’s significant correlation between Attrition & \n\nYearsWithCurrManager",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0041a77863352490141e03473f86483a40a1160 | 188,100 | ipynb | Jupyter Notebook | assignment2/ConvolutionalNetworks.ipynb | pranav-s/Stanford_CS234_CV_2017 | 9b0536812477dd0ea0e2dc4f063976a2e79148cc | [
"MIT"
] | null | null | null | assignment2/ConvolutionalNetworks.ipynb | pranav-s/Stanford_CS234_CV_2017 | 9b0536812477dd0ea0e2dc4f063976a2e79148cc | [
"MIT"
] | null | null | null | assignment2/ConvolutionalNetworks.ipynb | pranav-s/Stanford_CS234_CV_2017 | 9b0536812477dd0ea0e2dc4f063976a2e79148cc | [
"MIT"
] | null | null | null | 119.58042 | 132,028 | 0.846305 | [
[
[
"# Convolutional Networks\n\nSo far we have worked with deep fully-connected networks, using them to explore different optimization strategies and network architectures. Fully-connected networks are a good testbed for experimentation because they are very computationally efficient, but in practice all state-of-the-art results use convolutional networks instead.\n\nFirst you will implement several layer types that are used in convolutional networks. You will then use these layers to train a convolutional network on the CIFAR-10 dataset.",
"_____no_output_____"
]
],
[
[
"# As usual, a bit of setup\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom cs231n.classifiers.cnn import *\nfrom cs231n.data_utils import get_CIFAR10_data\nfrom cs231n.gradient_check import eval_numerical_gradient_array, eval_numerical_gradient\nfrom cs231n.layers import *\nfrom cs231n.fast_layers import *\nfrom cs231n.solver import Solver\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n# for auto-reloading external modules\n# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython\n%load_ext autoreload\n%autoreload 2\n\ndef rel_error(x, y):\n \"\"\" returns relative error \"\"\"\n return np.max(np.abs(x - y) / (np.maximum(1e-8, np.abs(x) + np.abs(y))))",
"_____no_output_____"
],
[
"# Load the (preprocessed) CIFAR10 data.\n\ndata = get_CIFAR10_data()\nfor k, v in data.items():\n print('%s: ' % k, v.shape)",
"X_train: (49000, 3, 32, 32)\ny_train: (49000,)\nX_val: (1000, 3, 32, 32)\ny_val: (1000,)\nX_test: (1000, 3, 32, 32)\ny_test: (1000,)\n"
]
],
[
[
"# Convolution: Naive forward pass\nThe core of a convolutional network is the convolution operation. In the file `cs231n/layers.py`, implement the forward pass for the convolution layer in the function `conv_forward_naive`. \n\nYou don't have to worry too much about efficiency at this point; just write the code in whatever way you find most clear.\n\nYou can test your implementation by running the following:",
"_____no_output_____"
]
],
[
[
"x_shape = (2, 3, 4, 4)\nw_shape = (3, 3, 4, 4)\nx = np.linspace(-0.1, 0.5, num=np.prod(x_shape)).reshape(x_shape)\nw = np.linspace(-0.2, 0.3, num=np.prod(w_shape)).reshape(w_shape)\nb = np.linspace(-0.1, 0.2, num=3)\n\nconv_param = {'stride': 2, 'pad': 1}\nout, _ = conv_forward_naive(x, w, b, conv_param)\ncorrect_out = np.array([[[[-0.08759809, -0.10987781],\n [-0.18387192, -0.2109216 ]],\n [[ 0.21027089, 0.21661097],\n [ 0.22847626, 0.23004637]],\n [[ 0.50813986, 0.54309974],\n [ 0.64082444, 0.67101435]]],\n [[[-0.98053589, -1.03143541],\n [-1.19128892, -1.24695841]],\n [[ 0.69108355, 0.66880383],\n [ 0.59480972, 0.56776003]],\n [[ 2.36270298, 2.36904306],\n [ 2.38090835, 2.38247847]]]])\n\n# Compare your output to ours; difference should be around e-8\nprint('Testing conv_forward_naive')\nprint('difference: ', rel_error(out, correct_out))",
"Testing conv_forward_naive\ndifference: 2.2121476417505994e-08\n"
],
[
"a = np.array([[[1,2,3], [3,2,5]],[[1,2,3], [3,2,5]],[[1,2,3], [3,2,5]]])\n#np.pad(a, 2, 'constant')\nimage_pad = np.array([np.pad(channel, 1 , 'constant') for channel in x])\nimage_pad.shape\nout.shape\n#w[0,:, 0:4, 0:4].shape",
"_____no_output_____"
]
],
[
[
"# Aside: Image processing via convolutions\n\nAs fun way to both check your implementation and gain a better understanding of the type of operation that convolutional layers can perform, we will set up an input containing two images and manually set up filters that perform common image processing operations (grayscale conversion and edge detection). The convolution forward pass will apply these operations to each of the input images. We can then visualize the results as a sanity check.",
"_____no_output_____"
],
[
"## Colab Users Only\n\nPlease execute the below cell to copy two cat images to the Colab VM.",
"_____no_output_____"
]
],
[
[
"# Colab users only!\n%mkdir -p cs231n/notebook_images\n%cd drive/My\\ Drive/$FOLDERNAME/cs231n\n%cp -r notebook_images/ /content/cs231n/\n%cd /content/",
"[Errno 2] No such file or directory: 'drive/My Drive/$FOLDERNAME/cs231n'\n/home/pranav/pCloudDrive/Atlanta/Coursework/Self-study/CS 231-Spring_2017/assignment2\ncp: cannot stat 'notebook_images/': No such file or directory\n[Errno 2] No such file or directory: '/content/'\n/home/pranav/pCloudDrive/Atlanta/Coursework/Self-study/CS 231-Spring_2017/assignment2\n"
],
[
"from imageio import imread\nfrom PIL import Image\n\nkitten = imread('cs231n/notebook_images/kitten.jpg')\npuppy = imread('cs231n/notebook_images/puppy.jpg')\n# kitten is wide, and puppy is already square\nd = kitten.shape[1] - kitten.shape[0]\nkitten_cropped = kitten[:, d//2:-d//2, :]\n\nimg_size = 200 # Make this smaller if it runs too slow\nresized_puppy = np.array(Image.fromarray(puppy).resize((img_size, img_size)))\nresized_kitten = np.array(Image.fromarray(kitten_cropped).resize((img_size, img_size)))\nx = np.zeros((2, 3, img_size, img_size))\nx[0, :, :, :] = resized_puppy.transpose((2, 0, 1))\nx[1, :, :, :] = resized_kitten.transpose((2, 0, 1))\n\n# Set up a convolutional weights holding 2 filters, each 3x3\nw = np.zeros((2, 3, 3, 3))\n\n# The first filter converts the image to grayscale.\n# Set up the red, green, and blue channels of the filter.\nw[0, 0, :, :] = [[0, 0, 0], [0, 0.3, 0], [0, 0, 0]]\nw[0, 1, :, :] = [[0, 0, 0], [0, 0.6, 0], [0, 0, 0]]\nw[0, 2, :, :] = [[0, 0, 0], [0, 0.1, 0], [0, 0, 0]]\n\n# Second filter detects horizontal edges in the blue channel.\nw[1, 2, :, :] = [[1, 2, 1], [0, 0, 0], [-1, -2, -1]]\n\n# Vector of biases. We don't need any bias for the grayscale\n# filter, but for the edge detection filter we want to add 128\n# to each output so that nothing is negative.\nb = np.array([0, 128])\n\n# Compute the result of convolving each input in x with each filter in w,\n# offsetting by b, and storing the results in out.\nout, _ = conv_forward_naive(x, w, b, {'stride': 1, 'pad': 1})\n\ndef imshow_no_ax(img, normalize=True):\n \"\"\" Tiny helper to show images as uint8 and remove axis labels \"\"\"\n if normalize:\n img_max, img_min = np.max(img), np.min(img)\n img = 255.0 * (img - img_min) / (img_max - img_min)\n plt.imshow(img.astype('uint8'))\n plt.gca().axis('off')\n\n# Show the original images and the results of the conv operation\nplt.subplot(2, 3, 1)\nimshow_no_ax(puppy, normalize=False)\nplt.title('Original image')\nplt.subplot(2, 3, 2)\nimshow_no_ax(out[0, 0])\nplt.title('Grayscale')\nplt.subplot(2, 3, 3)\nimshow_no_ax(out[0, 1])\nplt.title('Edges')\nplt.subplot(2, 3, 4)\nimshow_no_ax(kitten_cropped, normalize=False)\nplt.subplot(2, 3, 5)\nimshow_no_ax(out[1, 0])\nplt.subplot(2, 3, 6)\nimshow_no_ax(out[1, 1])\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Convolution: Naive backward pass\nImplement the backward pass for the convolution operation in the function `conv_backward_naive` in the file `cs231n/layers.py`. Again, you don't need to worry too much about computational efficiency.\n\nWhen you are done, run the following to check your backward pass with a numeric gradient check.",
"_____no_output_____"
]
],
[
[
"np.random.seed(231)\nx = np.random.randn(4, 3, 5, 5)\nw = np.random.randn(2, 3, 3, 3)\nb = np.random.randn(2,)\ndout = np.random.randn(4, 2, 5, 5)\nconv_param = {'stride': 2, 'pad': 3}\n\nout, cache = conv_forward_naive(x, w, b, conv_param)\ndx, dw, db = conv_backward_naive(dout, cache)\n\ndx_num = eval_numerical_gradient_array(lambda x: conv_forward_naive(x, w, b, conv_param)[0], x, dout)\ndw_num = eval_numerical_gradient_array(lambda w: conv_forward_naive(x, w, b, conv_param)[0], w, dout)\ndb_num = eval_numerical_gradient_array(lambda b: conv_forward_naive(x, w, b, conv_param)[0], b, dout)\n\n# Your errors should be around e-8 or less.\nprint('Testing conv_backward_naive function')\nprint('dx error: ', rel_error(dx, dx_num))\nprint('dw error: ', rel_error(dw, dw_num))\nprint('db error: ', rel_error(db, db_num))",
"Testing conv_backward_naive function\ndx error: 2.581952248938191e-09\ndw error: 3.4015113317197913e-10\ndb error: 8.870685436057798e-11\n"
],
[
"print(dw_num)\nprint(dw)",
"[[[[ 3.74083229 -1.11736932 5.65008475]\n [-19.11962989 5.65406037 4.53124725]\n [ -3.6329338 3.76169087 7.20472087]]\n\n [[ 7.03395312 -4.5727834 -7.8451423 ]\n [ 5.66691388 -12.92959222 -0.40313598]\n [ 5.28682252 11.21425667 -3.41223078]]\n\n [[ 9.39415069 -22.2121015 -16.20336304]\n [-14.39395302 -15.52157928 -8.1869118 ]\n [ 2.5098062 -2.08364313 -1.90675446]]]\n\n\n [[[ -9.06570861 11.87244296 -7.41391816]\n [ -0.12837405 -7.23888876 9.17635209]\n [ 4.14548712 23.18841142 -9.49222648]]\n\n [[ 2.58171801 -1.78163112 -1.10845452]\n [-13.76007062 3.91269497 -6.56275002]\n [ 3.27507715 -4.38790371 10.09310656]]\n\n [[-14.34939674 -2.80296041 -3.72368111]\n [ 0.65154294 24.28001799 -10.36012214]\n [ -3.16236407 1.52530603 3.68371982]]]]\n[[[[ 7.74083229 2.88263068 9.65008475]\n [-15.11962989 9.65406037 8.53124725]\n [ 0.3670662 7.76169087 11.20472087]]\n\n [[ 11.03395312 -0.5727834 -3.8451423 ]\n [ 9.66691388 -8.92959222 3.59686402]\n [ 9.28682252 15.21425667 0.58776922]]\n\n [[ 13.39415069 -18.2121015 -12.20336304]\n [-10.39395302 -11.52157928 -4.1869118 ]\n [ 6.5098062 1.91635687 2.09324554]]]\n\n\n [[[ -5.06570861 15.87244296 -3.41391816]\n [ 3.87162595 -3.23888876 13.17635209]\n [ 8.14548712 27.18841142 -5.49222648]]\n\n [[ 6.58171801 2.21836888 2.89154548]\n [ -9.76007062 7.91269497 -2.56275002]\n [ 7.27507715 -0.38790371 14.09310656]]\n\n [[-10.34939674 1.19703959 0.27631889]\n [ 4.65154294 28.28001799 -6.36012214]\n [ 0.83763593 5.52530603 7.68371982]]]]\n"
],
[
"t = np.array([[1,2], [3,4]])\nnp.rot90(t, k=2)",
"_____no_output_____"
]
],
[
[
"# Max-Pooling: Naive forward\nImplement the forward pass for the max-pooling operation in the function `max_pool_forward_naive` in the file `cs231n/layers.py`. Again, don't worry too much about computational efficiency.\n\nCheck your implementation by running the following:",
"_____no_output_____"
]
],
[
[
"x_shape = (2, 3, 4, 4)\nx = np.linspace(-0.3, 0.4, num=np.prod(x_shape)).reshape(x_shape)\npool_param = {'pool_width': 2, 'pool_height': 2, 'stride': 2}\n\nout, _ = max_pool_forward_naive(x, pool_param)\n\ncorrect_out = np.array([[[[-0.26315789, -0.24842105],\n [-0.20421053, -0.18947368]],\n [[-0.14526316, -0.13052632],\n [-0.08631579, -0.07157895]],\n [[-0.02736842, -0.01263158],\n [ 0.03157895, 0.04631579]]],\n [[[ 0.09052632, 0.10526316],\n [ 0.14947368, 0.16421053]],\n [[ 0.20842105, 0.22315789],\n [ 0.26736842, 0.28210526]],\n [[ 0.32631579, 0.34105263],\n [ 0.38526316, 0.4 ]]]])\n\n# Compare your output with ours. Difference should be on the order of e-8.\nprint('Testing max_pool_forward_naive function:')\nprint('difference: ', rel_error(out, correct_out))",
"Testing max_pool_forward_naive function:\ndifference: 4.1666665157267834e-08\n"
]
],
[
[
"# Max-Pooling: Naive backward\nImplement the backward pass for the max-pooling operation in the function `max_pool_backward_naive` in the file `cs231n/layers.py`. You don't need to worry about computational efficiency.\n\nCheck your implementation with numeric gradient checking by running the following:",
"_____no_output_____"
]
],
[
[
"np.random.seed(231)\nx = np.random.randn(3, 2, 8, 8)\ndout = np.random.randn(3, 2, 4, 4)\npool_param = {'pool_height': 2, 'pool_width': 2, 'stride': 2}\n\ndx_num = eval_numerical_gradient_array(lambda x: max_pool_forward_naive(x, pool_param)[0], x, dout)\n\nout, cache = max_pool_forward_naive(x, pool_param)\ndx = max_pool_backward_naive(dout, cache)\n\n# Your error should be on the order of e-12\nprint('Testing max_pool_backward_naive function:')\nprint('dx error: ', rel_error(dx, dx_num))",
"Testing max_pool_backward_naive function:\ndx error: 3.27562514223145e-12\n"
]
],
[
[
"# Fast layers\n\nMaking convolution and pooling layers fast can be challenging. To spare you the pain, we've provided fast implementations of the forward and backward passes for convolution and pooling layers in the file `cs231n/fast_layers.py`.",
"_____no_output_____"
],
[
"The fast convolution implementation depends on a Cython extension; to compile it either execute the local development cell (option A) if you are developing locally, or the Colab cell (option B) if you are running this assignment in Colab.\n\n---\n\n**Very Important, Please Read**. For **both** option A and B, you have to **restart** the notebook after compiling the cython extension. In Colab, please save the notebook `File -> Save`, then click `Runtime -> Restart Runtime -> Yes`. This will restart the kernel which means local variables will be lost. Just re-execute the cells from top to bottom and skip the cell below as you only need to run it once for the compilation step.\n\n---",
"_____no_output_____"
],
[
"## Option A: Local Development\n\nGo to the cs231n directory and execute the following in your terminal:\n\n```bash\npython setup.py build_ext --inplace\n```",
"_____no_output_____"
],
[
"## Option B: Colab\n\nExecute the cell below only only **ONCE**.",
"_____no_output_____"
]
],
[
[
"%cd drive/My\\ Drive/$FOLDERNAME/cs231n/\n!python setup.py build_ext --inplace",
"_____no_output_____"
]
],
[
[
"The API for the fast versions of the convolution and pooling layers is exactly the same as the naive versions that you implemented above: the forward pass receives data, weights, and parameters and produces outputs and a cache object; the backward pass recieves upstream derivatives and the cache object and produces gradients with respect to the data and weights.\n\n**NOTE:** The fast implementation for pooling will only perform optimally if the pooling regions are non-overlapping and tile the input. If these conditions are not met then the fast pooling implementation will not be much faster than the naive implementation.\n\nYou can compare the performance of the naive and fast versions of these layers by running the following:",
"_____no_output_____"
]
],
[
[
"# Rel errors should be around e-9 or less\nfrom cs231n.fast_layers import conv_forward_fast, conv_backward_fast\nfrom time import time\n%load_ext autoreload\n%autoreload 2\nnp.random.seed(231)\nx = np.random.randn(100, 3, 31, 31)\nw = np.random.randn(25, 3, 3, 3)\nb = np.random.randn(25,)\ndout = np.random.randn(100, 25, 16, 16)\nconv_param = {'stride': 2, 'pad': 1}\n\nt0 = time()\nout_naive, cache_naive = conv_forward_naive(x, w, b, conv_param)\nt1 = time()\nout_fast, cache_fast = conv_forward_fast(x, w, b, conv_param)\nt2 = time()\n\nprint('Testing conv_forward_fast:')\nprint('Naive: %fs' % (t1 - t0))\nprint('Fast: %fs' % (t2 - t1))\nprint('Speedup: %fx' % ((t1 - t0) / (t2 - t1)))\nprint('Difference: ', rel_error(out_naive, out_fast))\n\nt0 = time()\n# dx_naive, dw_naive, db_naive = conv_backward_naive(dout, cache_naive)\nt1 = time()\ndx_fast, dw_fast, db_fast = conv_backward_fast(dout, cache_fast)\nt2 = time()\n\nprint('\\nTesting conv_backward_fast:')\nprint('Naive: %fs' % (t1 - t0))\nprint('Fast: %fs' % (t2 - t1))\nprint('Speedup: %fx' % ((t1 - t0) / (t2 - t1)))\n# print('dx difference: ', rel_error(dx_naive, dx_fast))\n# print('dw difference: ', rel_error(dw_naive, dw_fast))\n# print('db difference: ', rel_error(db_naive, db_fast))",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\nTesting conv_forward_fast:\nNaive: 3.358959s\nFast: 0.004483s\nSpeedup: 749.188726x\nDifference: 4.926407851494105e-11\n\nTesting conv_backward_fast:\nNaive: 0.000029s\nFast: 0.007967s\nSpeedup: 0.003651x\n"
],
[
"# Relative errors should be close to 0.0\nfrom cs231n.fast_layers import max_pool_forward_fast, max_pool_backward_fast\nnp.random.seed(231)\nx = np.random.randn(100, 3, 32, 32)\ndout = np.random.randn(100, 3, 16, 16)\npool_param = {'pool_height': 2, 'pool_width': 2, 'stride': 2}\n\nt0 = time()\nout_naive, cache_naive = max_pool_forward_naive(x, pool_param)\nt1 = time()\nout_fast, cache_fast = max_pool_forward_fast(x, pool_param)\nt2 = time()\n\nprint('Testing pool_forward_fast:')\nprint('Naive: %fs' % (t1 - t0))\nprint('fast: %fs' % (t2 - t1))\nprint('speedup: %fx' % ((t1 - t0) / (t2 - t1)))\nprint('difference: ', rel_error(out_naive, out_fast))\n\nt0 = time()\ndx_naive = max_pool_backward_naive(dout, cache_naive)\nt1 = time()\ndx_fast = max_pool_backward_fast(dout, cache_fast)\nt2 = time()\n\nprint('\\nTesting pool_backward_fast:')\nprint('Naive: %fs' % (t1 - t0))\nprint('fast: %fs' % (t2 - t1))\nprint('speedup: %fx' % ((t1 - t0) / (t2 - t1)))\nprint('dx difference: ', rel_error(dx_naive, dx_fast))",
"Testing pool_forward_fast:\nNaive: 0.248409s\nfast: 0.001589s\nspeedup: 156.347839x\ndifference: 0.0\n\nTesting pool_backward_fast:\nNaive: 0.323171s\nfast: 0.007276s\nspeedup: 44.415689x\ndx difference: 0.0\n"
]
],
[
[
"# Convolutional \"sandwich\" layers\nPreviously we introduced the concept of \"sandwich\" layers that combine multiple operations into commonly used patterns. In the file `cs231n/layer_utils.py` you will find sandwich layers that implement a few commonly used patterns for convolutional networks. Run the cells below to sanity check they're working.",
"_____no_output_____"
]
],
[
[
"from cs231n.layer_utils import conv_relu_pool_forward, conv_relu_pool_backward\nnp.random.seed(231)\nx = np.random.randn(2, 3, 16, 16)\nw = np.random.randn(3, 3, 3, 3)\nb = np.random.randn(3,)\ndout = np.random.randn(2, 3, 8, 8)\nconv_param = {'stride': 1, 'pad': 1}\npool_param = {'pool_height': 2, 'pool_width': 2, 'stride': 2}\n\nout, cache = conv_relu_pool_forward(x, w, b, conv_param, pool_param)\ndx, dw, db = conv_relu_pool_backward(dout, cache)\n\ndx_num = eval_numerical_gradient_array(lambda x: conv_relu_pool_forward(x, w, b, conv_param, pool_param)[0], x, dout)\ndw_num = eval_numerical_gradient_array(lambda w: conv_relu_pool_forward(x, w, b, conv_param, pool_param)[0], w, dout)\ndb_num = eval_numerical_gradient_array(lambda b: conv_relu_pool_forward(x, w, b, conv_param, pool_param)[0], b, dout)\n\n# Relative errors should be around e-8 or less\nprint('Testing conv_relu_pool')\nprint('dx error: ', rel_error(dx_num, dx))\nprint('dw error: ', rel_error(dw_num, dw))\nprint('db error: ', rel_error(db_num, db))",
"Testing conv_relu_pool\ndx error: 9.591132621921372e-09\ndw error: 5.802391137330214e-09\ndb error: 1.0146343411762047e-09\n"
],
[
"from cs231n.layer_utils import conv_relu_forward, conv_relu_backward\nnp.random.seed(231)\nx = np.random.randn(2, 3, 8, 8)\nw = np.random.randn(3, 3, 3, 3)\nb = np.random.randn(3,)\ndout = np.random.randn(2, 3, 8, 8)\nconv_param = {'stride': 1, 'pad': 1}\n\nout, cache = conv_relu_forward(x, w, b, conv_param)\ndx, dw, db = conv_relu_backward(dout, cache)\n\ndx_num = eval_numerical_gradient_array(lambda x: conv_relu_forward(x, w, b, conv_param)[0], x, dout)\ndw_num = eval_numerical_gradient_array(lambda w: conv_relu_forward(x, w, b, conv_param)[0], w, dout)\ndb_num = eval_numerical_gradient_array(lambda b: conv_relu_forward(x, w, b, conv_param)[0], b, dout)\n\n# Relative errors should be around e-8 or less\nprint('Testing conv_relu:')\nprint('dx error: ', rel_error(dx_num, dx))\nprint('dw error: ', rel_error(dw_num, dw))\nprint('db error: ', rel_error(db_num, db))",
"Testing conv_relu:\ndx error: 1.5218619980349303e-09\ndw error: 2.702022646099404e-10\ndb error: 1.451272393591721e-10\n"
]
],
[
[
"# Three-layer ConvNet\nNow that you have implemented all the necessary layers, we can put them together into a simple convolutional network.\n\nOpen the file `cs231n/classifiers/cnn.py` and complete the implementation of the `ThreeLayerConvNet` class. Remember you can use the fast/sandwich layers (already imported for you) in your implementation. Run the following cells to help you debug:",
"_____no_output_____"
],
[
"## Sanity check loss\nAfter you build a new network, one of the first things you should do is sanity check the loss. When we use the softmax loss, we expect the loss for random weights (and no regularization) to be about `log(C)` for `C` classes. When we add regularization the loss should go up slightly.",
"_____no_output_____"
]
],
[
[
"model = ThreeLayerConvNet()\n\nN = 50\nX = np.random.randn(N, 3, 32, 32)\ny = np.random.randint(10, size=N)\n\nloss, grads = model.loss(X, y)\nprint('Initial loss (no regularization): ', loss)\n\nmodel.reg = 0.5\nloss, grads = model.loss(X, y)\nprint('Initial loss (with regularization): ', loss)",
"Initial loss (no regularization): 2.3025850635890874\nInitial loss (with regularization): 2.508599728507643\n"
]
],
[
[
"## Gradient check\nAfter the loss looks reasonable, use numeric gradient checking to make sure that your backward pass is correct. When you use numeric gradient checking you should use a small amount of artifical data and a small number of neurons at each layer. Note: correct implementations may still have relative errors up to the order of e-2.",
"_____no_output_____"
]
],
[
[
"num_inputs = 2\ninput_dim = (3, 16, 16)\nreg = 0.0\nnum_classes = 10\nnp.random.seed(231)\nX = np.random.randn(num_inputs, *input_dim)\ny = np.random.randint(num_classes, size=num_inputs)\n\nmodel = ThreeLayerConvNet(num_filters=3, filter_size=3,\n input_dim=input_dim, hidden_dim=7,\n dtype=np.float64, reg=0.5)\nloss, grads = model.loss(X, y)\n# Errors should be small, but correct implementations may have\n# relative errors up to the order of e-2\nfor param_name in sorted(grads):\n f = lambda _: model.loss(X, y)[0]\n param_grad_num = eval_numerical_gradient(f, model.params[param_name], verbose=False, h=1e-6)\n e = rel_error(param_grad_num, grads[param_name])\n print('%s max relative error: %e' % (param_name, rel_error(param_grad_num, grads[param_name])))",
"W1 max relative error: 1.103635e-05\nW2 max relative error: 1.521379e-04\nW3 max relative error: 1.763147e-05\nb1 max relative error: 3.477652e-05\nb2 max relative error: 2.516375e-03\nb3 max relative error: 7.945660e-10\n"
]
],
[
[
"## Overfit small data\nA nice trick is to train your model with just a few training samples. You should be able to overfit small datasets, which will result in very high training accuracy and comparatively low validation accuracy.",
"_____no_output_____"
]
],
[
[
"np.random.seed(231)\n\nnum_train = 100\nsmall_data = {\n 'X_train': data['X_train'][:num_train],\n 'y_train': data['y_train'][:num_train],\n 'X_val': data['X_val'],\n 'y_val': data['y_val'],\n}\n\nmodel = ThreeLayerConvNet(weight_scale=1e-2)\n\nsolver = Solver(model, small_data,\n num_epochs=15, batch_size=50,\n update_rule='adam',\n optim_config={\n 'learning_rate': 1e-3,\n },\n verbose=True, print_every=1)\nsolver.train()",
"_____no_output_____"
],
[
"# Print final training accuracy\nprint(\n \"Small data training accuracy:\",\n solver.check_accuracy(small_data['X_train'], small_data['y_train'])\n)",
"_____no_output_____"
],
[
"# Print final validation accuracy\nprint(\n \"Small data validation accuracy:\",\n solver.check_accuracy(small_data['X_val'], small_data['y_val'])\n)",
"_____no_output_____"
]
],
[
[
"Plotting the loss, training accuracy, and validation accuracy should show clear overfitting:",
"_____no_output_____"
]
],
[
[
"plt.subplot(2, 1, 1)\nplt.plot(solver.loss_history, 'o')\nplt.xlabel('iteration')\nplt.ylabel('loss')\n\nplt.subplot(2, 1, 2)\nplt.plot(solver.train_acc_history, '-o')\nplt.plot(solver.val_acc_history, '-o')\nplt.legend(['train', 'val'], loc='upper left')\nplt.xlabel('epoch')\nplt.ylabel('accuracy')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Train the net\nBy training the three-layer convolutional network for one epoch, you should achieve greater than 40% accuracy on the training set:",
"_____no_output_____"
]
],
[
[
"model = ThreeLayerConvNet(weight_scale=0.001, hidden_dim=500, reg=0.001)\n\nsolver = Solver(model, data,\n num_epochs=1, batch_size=50,\n update_rule='adam',\n optim_config={\n 'learning_rate': 1e-3,\n },\n verbose=True, print_every=20)\nsolver.train()",
"_____no_output_____"
],
[
"# Print final training accuracy\nprint(\n \"Full data training accuracy:\",\n solver.check_accuracy(small_data['X_train'], small_data['y_train'])\n)",
"_____no_output_____"
],
[
"# Print final validation accuracy\nprint(\n \"Full data validation accuracy:\",\n solver.check_accuracy(data['X_val'], data['y_val'])\n)",
"_____no_output_____"
]
],
[
[
"## Visualize Filters\nYou can visualize the first-layer convolutional filters from the trained network by running the following:",
"_____no_output_____"
]
],
[
[
"from cs231n.vis_utils import visualize_grid\n\ngrid = visualize_grid(model.params['W1'].transpose(0, 2, 3, 1))\nplt.imshow(grid.astype('uint8'))\nplt.axis('off')\nplt.gcf().set_size_inches(5, 5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Spatial Batch Normalization\nWe already saw that batch normalization is a very useful technique for training deep fully-connected networks. As proposed in the original paper (link in `BatchNormalization.ipynb`), batch normalization can also be used for convolutional networks, but we need to tweak it a bit; the modification will be called \"spatial batch normalization.\"\n\nNormally batch-normalization accepts inputs of shape `(N, D)` and produces outputs of shape `(N, D)`, where we normalize across the minibatch dimension `N`. For data coming from convolutional layers, batch normalization needs to accept inputs of shape `(N, C, H, W)` and produce outputs of shape `(N, C, H, W)` where the `N` dimension gives the minibatch size and the `(H, W)` dimensions give the spatial size of the feature map.\n\nIf the feature map was produced using convolutions, then we expect every feature channel's statistics e.g. mean, variance to be relatively consistent both between different images, and different locations within the same image -- after all, every feature channel is produced by the same convolutional filter! Therefore spatial batch normalization computes a mean and variance for each of the `C` feature channels by computing statistics over the minibatch dimension `N` as well the spatial dimensions `H` and `W`.\n\n\n[1] [Sergey Ioffe and Christian Szegedy, \"Batch Normalization: Accelerating Deep Network Training by Reducing\nInternal Covariate Shift\", ICML 2015.](https://arxiv.org/abs/1502.03167)",
"_____no_output_____"
],
[
"## Spatial batch normalization: forward\n\nIn the file `cs231n/layers.py`, implement the forward pass for spatial batch normalization in the function `spatial_batchnorm_forward`. Check your implementation by running the following:",
"_____no_output_____"
]
],
[
[
"np.random.seed(231)\n# Check the training-time forward pass by checking means and variances\n# of features both before and after spatial batch normalization\n\nN, C, H, W = 2, 3, 4, 5\nx = 4 * np.random.randn(N, C, H, W) + 10\n\nprint('Before spatial batch normalization:')\nprint(' Shape: ', x.shape)\nprint(' Means: ', x.mean(axis=(0, 2, 3)))\nprint(' Stds: ', x.std(axis=(0, 2, 3)))\n\n# Means should be close to zero and stds close to one\ngamma, beta = np.ones(C), np.zeros(C)\nbn_param = {'mode': 'train'}\nout, _ = spatial_batchnorm_forward(x, gamma, beta, bn_param)\nprint('After spatial batch normalization:')\nprint(' Shape: ', out.shape)\nprint(' Means: ', out.mean(axis=(0, 2, 3)))\nprint(' Stds: ', out.std(axis=(0, 2, 3)))\n\n# Means should be close to beta and stds close to gamma\ngamma, beta = np.asarray([3, 4, 5]), np.asarray([6, 7, 8])\nout, _ = spatial_batchnorm_forward(x, gamma, beta, bn_param)\nprint('After spatial batch normalization (nontrivial gamma, beta):')\nprint(' Shape: ', out.shape)\nprint(' Means: ', out.mean(axis=(0, 2, 3)))\nprint(' Stds: ', out.std(axis=(0, 2, 3)))",
"Before spatial batch normalization:\n Shape: (2, 3, 4, 5)\n Means: [9.33463814 8.90909116 9.11056338]\n Stds: [3.61447857 3.19347686 3.5168142 ]\nAfter spatial batch normalization:\n Shape: (2, 3, 4, 5)\n Means: [0.00000000e+00 3.10862447e-16 9.43689571e-17]\n Stds: [0.99998977 0.99987472 0.99999591]\nAfter spatial batch normalization (nontrivial gamma, beta):\n Shape: (2, 3, 4, 5)\n Means: [6. 7. 8.]\n Stds: [2.99996931 3.99949889 4.99997957]\n"
],
[
"np.random.seed(231)\n# Check the test-time forward pass by running the training-time\n# forward pass many times to warm up the running averages, and then\n# checking the means and variances of activations after a test-time\n# forward pass.\nN, C, H, W = 10, 4, 11, 12\n\nbn_param = {'mode': 'train'}\ngamma = np.ones(C)\nbeta = np.zeros(C)\nfor t in range(50):\n x = 2.3 * np.random.randn(N, C, H, W) + 13\n spatial_batchnorm_forward(x, gamma, beta, bn_param)\nbn_param['mode'] = 'test'\nx = 2.3 * np.random.randn(N, C, H, W) + 13\na_norm, _ = spatial_batchnorm_forward(x, gamma, beta, bn_param)\n\n# Means should be close to zero and stds close to one, but will be\n# noisier than training-time forward passes.\nprint('After spatial batch normalization (test-time):')\nprint(' means: ', a_norm.mean(axis=(0, 2, 3)))\nprint(' stds: ', a_norm.std(axis=(0, 2, 3)))",
"After spatial batch normalization (test-time):\n means: [-0.08446363 0.08091916 0.06055194 0.04564399]\n stds: [1.0241906 1.09568294 1.0903571 1.0684257 ]\n"
]
],
[
[
"## Spatial batch normalization: backward\nIn the file `cs231n/layers.py`, implement the backward pass for spatial batch normalization in the function `spatial_batchnorm_backward`. Run the following to check your implementation using a numeric gradient check:",
"_____no_output_____"
]
],
[
[
"np.random.seed(231)\nN, C, H, W = 2, 3, 4, 5\nx = 5 * np.random.randn(N, C, H, W) + 12\ngamma = np.random.randn(C)\nbeta = np.random.randn(C)\ndout = np.random.randn(N, C, H, W)\n\nbn_param = {'mode': 'train'}\nfx = lambda x: spatial_batchnorm_forward(x, gamma, beta, bn_param)[0]\nfg = lambda a: spatial_batchnorm_forward(x, gamma, beta, bn_param)[0]\nfb = lambda b: spatial_batchnorm_forward(x, gamma, beta, bn_param)[0]\n\ndx_num = eval_numerical_gradient_array(fx, x, dout)\nda_num = eval_numerical_gradient_array(fg, gamma, dout)\ndb_num = eval_numerical_gradient_array(fb, beta, dout)\n\n#You should expect errors of magnitudes between 1e-12~1e-06\n_, cache = spatial_batchnorm_forward(x, gamma, beta, bn_param)\ndx, dgamma, dbeta = spatial_batchnorm_backward(dout, cache)\nprint('dx error: ', rel_error(dx_num, dx))\nprint('dgamma error: ', rel_error(da_num, dgamma))\nprint('dbeta error: ', rel_error(db_num, dbeta))",
"dx error: 0.0011090161391138292\ndgamma error: 4.5988216115954125e-11\ndbeta error: 3.2755517433052766e-12\n"
],
[
"print(dx[0])\nprint(dx_num[0])\nprint(rel_error(dx[0], dx_num[0]))",
"[[[ 4.82500057e-08 -6.18616346e-07 3.49900510e-08 -1.01656172e-06\n 2.21274578e-04]\n [-8.18296259e-07 1.06936237e-07 2.56172679e-08 6.08105008e-08\n -7.21749724e-06]\n [-1.88721588e-05 3.50657245e-07 -3.24214426e-06 -9.50347872e-08\n 3.30202440e-07]\n [-4.69853077e-06 -3.05069739e-07 9.78361879e-08 7.28839098e-06\n -4.55892654e-08]]\n\n [[-1.04277935e-07 9.63218452e-07 1.98862366e-06 3.38830371e-06\n 1.56879316e-06]\n [-1.76798576e-06 -2.04511247e-04 1.24032782e-02 -8.19890551e-10\n 8.00572597e-06]\n [-1.97819314e-08 -2.21420815e-07 -2.18365091e-07 5.12938128e-07\n 5.80098461e-06]\n [-7.32695890e-08 1.74940725e-08 -5.10295706e-07 2.65071171e-05\n 6.13496551e-08]]\n\n [[ 4.10499437e-09 -1.64792199e-09 7.77772208e-08 -3.98336745e-08\n -5.19554157e-07]\n [ 8.93598739e-10 1.54241596e-09 -6.17399690e-10 5.98332851e-11\n 1.54458330e-09]\n [-1.78912331e-07 4.76659981e-10 1.29507309e-08 1.66509831e-09\n 1.57005474e-09]\n [ 5.76918608e-10 -5.78221607e-10 1.91686211e-09 7.21027908e-08\n -7.31198445e-11]]]\n[[[ 4.82429917e-08 -6.18613071e-07 3.49810343e-08 -1.01655161e-06\n 2.21274577e-04]\n [-8.18296101e-07 1.06943155e-07 2.56248467e-08 6.08208895e-08\n -7.21751834e-06]\n [-1.88721444e-05 3.50657095e-07 -3.24213026e-06 -9.50347287e-08\n 3.30190632e-07]\n [-4.69853074e-06 -3.05076200e-07 9.78253870e-08 7.28839423e-06\n -4.55828488e-08]]\n\n [[-1.04280277e-07 9.63219277e-07 1.98862054e-06 3.38830579e-06\n 1.56879241e-06]\n [-1.76798531e-06 -2.04511240e-04 1.24032784e-02 -8.19870920e-10\n 8.00571709e-06]\n [-1.97798693e-08 -2.21423050e-07 -2.18359769e-07 5.12933335e-07\n 5.80098067e-06]\n [-7.32873779e-08 1.74943463e-08 -5.10310657e-07 2.65071114e-05\n 6.13411225e-08]]\n\n [[ 4.10535500e-09 -1.64264801e-09 7.77871231e-08 -3.98317671e-08\n -5.19546842e-07]\n [ 8.99421228e-10 1.53625905e-09 -6.14967580e-10 5.86159573e-11\n 1.54866956e-09]\n [-1.78944574e-07 4.76763898e-10 1.29497212e-08 1.66313198e-09\n 1.57114321e-09]\n [ 5.78678420e-10 -5.89311768e-10 1.91779211e-09 7.20991748e-08\n -7.49812329e-11]]]\n0.0011090161282718075\n"
]
],
[
[
"# Group Normalization\nIn the previous notebook, we mentioned that Layer Normalization is an alternative normalization technique that mitigates the batch size limitations of Batch Normalization. However, as the authors of [2] observed, Layer Normalization does not perform as well as Batch Normalization when used with Convolutional Layers:\n\n>With fully connected layers, all the hidden units in a layer tend to make similar contributions to the final prediction, and re-centering and rescaling the summed inputs to a layer works well. However, the assumption of similar contributions is no longer true for convolutional neural networks. The large number of the hidden units whose\nreceptive fields lie near the boundary of the image are rarely turned on and thus have very different\nstatistics from the rest of the hidden units within the same layer.\n\nThe authors of [3] propose an intermediary technique. In contrast to Layer Normalization, where you normalize over the entire feature per-datapoint, they suggest a consistent splitting of each per-datapoint feature into G groups, and a per-group per-datapoint normalization instead. \n\n<p align=\"center\">\n<img src=\"https://raw.githubusercontent.com/cs231n/cs231n.github.io/master/assets/a2/normalization.png\">\n</p>\n<center>Visual comparison of the normalization techniques discussed so far (image edited from [3])</center>\n\nEven though an assumption of equal contribution is still being made within each group, the authors hypothesize that this is not as problematic, as innate grouping arises within features for visual recognition. One example they use to illustrate this is that many high-performance handcrafted features in traditional Computer Vision have terms that are explicitly grouped together. Take for example Histogram of Oriented Gradients [4]-- after computing histograms per spatially local block, each per-block histogram is normalized before being concatenated together to form the final feature vector.\n\nYou will now implement Group Normalization. Note that this normalization technique that you are to implement in the following cells was introduced and published to ECCV just in 2018 -- this truly is still an ongoing and excitingly active field of research!\n\n[2] [Ba, Jimmy Lei, Jamie Ryan Kiros, and Geoffrey E. Hinton. \"Layer Normalization.\" stat 1050 (2016): 21.](https://arxiv.org/pdf/1607.06450.pdf)\n\n\n[3] [Wu, Yuxin, and Kaiming He. \"Group Normalization.\" arXiv preprint arXiv:1803.08494 (2018).](https://arxiv.org/abs/1803.08494)\n\n\n[4] [N. Dalal and B. Triggs. Histograms of oriented gradients for\nhuman detection. In Computer Vision and Pattern Recognition\n(CVPR), 2005.](https://ieeexplore.ieee.org/abstract/document/1467360/)",
"_____no_output_____"
],
[
"## Group normalization: forward\n\nIn the file `cs231n/layers.py`, implement the forward pass for group normalization in the function `spatial_groupnorm_forward`. Check your implementation by running the following:",
"_____no_output_____"
]
],
[
[
"np.random.seed(231)\n# Check the training-time forward pass by checking means and variances\n# of features both before and after spatial batch normalization\n\nN, C, H, W = 2, 6, 4, 5\nG = 2\nx = 4 * np.random.randn(N, C, H, W) + 10\nx_g = x.reshape((N*G,-1))\nprint('Before spatial group normalization:')\nprint(' Shape: ', x.shape)\nprint(' Means: ', x_g.mean(axis=1))\nprint(' Stds: ', x_g.std(axis=1))\n\n# Means should be close to zero and stds close to one\ngamma, beta = 2*np.ones(C), np.zeros(C)\nbn_param = {'mode': 'train'}\n\nout, _ = spatial_groupnorm_quick_forward(x, gamma, beta, G, bn_param)\nout_g = out.reshape((N*G,-1))\nprint('After spatial group normalization:')\nprint(' Shape: ', out.shape)\nprint(' Means: ', out_g.mean(axis=1))\nprint(' Stds: ', out_g.std(axis=1))",
"Before spatial group normalization:\n Shape: (2, 6, 4, 5)\n Means: [9.72505327 8.51114185 8.9147544 9.43448077]\n Stds: [3.67070958 3.09892597 4.27043622 3.97521327]\nAfter spatial group normalization:\n Shape: (2, 6, 4, 5)\n Means: [-4.29286236e-16 1.05101113e-15 5.31056680e-16 -6.77236045e-16]\n Stds: [1.99999926 1.99999896 1.99999945 1.99999937]\n"
],
[
"np.vstack(list([np.hstack([[g]*H*W for g in gamma])])*N).shape",
"_____no_output_____"
],
[
"p = np.zeros((3,4))\nprint(p)\nq = np.hsplit(p, 2)\nprint(q)\nnp.hstack(q)",
"[[0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]]\n[array([[0., 0.],\n [0., 0.],\n [0., 0.]]), array([[0., 0.],\n [0., 0.],\n [0., 0.]])]\n"
],
[
"print(np.arange(36).reshape((6,6)).reshape((18,-1)))\nprint(np.arange(36).reshape((6,6)))\nprint(np.arange(36).reshape((6,6)).reshape((18,-1)).reshape((6, -1)))",
"[[ 0 1]\n [ 2 3]\n [ 4 5]\n [ 6 7]\n [ 8 9]\n [10 11]\n [12 13]\n [14 15]\n [16 17]\n [18 19]\n [20 21]\n [22 23]\n [24 25]\n [26 27]\n [28 29]\n [30 31]\n [32 33]\n [34 35]]\n[[ 0 1 2 3 4 5]\n [ 6 7 8 9 10 11]\n [12 13 14 15 16 17]\n [18 19 20 21 22 23]\n [24 25 26 27 28 29]\n [30 31 32 33 34 35]]\n[[ 0 1 2 3 4 5]\n [ 6 7 8 9 10 11]\n [12 13 14 15 16 17]\n [18 19 20 21 22 23]\n [24 25 26 27 28 29]\n [30 31 32 33 34 35]]\n"
]
],
[
[
"## Spatial group normalization: backward\nIn the file `cs231n/layers.py`, implement the backward pass for spatial batch normalization in the function `spatial_groupnorm_backward`. Run the following to check your implementation using a numeric gradient check:",
"_____no_output_____"
]
],
[
[
"np.random.seed(231)\nN, C, H, W = 2, 6, 4, 5\nG = 2\nx = 5 * np.random.randn(N, C, H, W) + 12\ngamma = np.random.randn(C)\nbeta = np.random.randn(C)\ndout = np.random.randn(N, C, H, W)\n\ngn_param = {}\nfx = lambda x: spatial_groupnorm_quick_forward(x, gamma, beta, G, gn_param)[0]\nfg = lambda a: spatial_groupnorm_quick_forward(x, gamma, beta, G, gn_param)[0]\nfb = lambda b: spatial_groupnorm_quick_forward(x, gamma, beta, G, gn_param)[0]\n\ndx_num = eval_numerical_gradient_array(fx, x, dout)\nda_num = eval_numerical_gradient_array(fg, gamma, dout)\ndb_num = eval_numerical_gradient_array(fb, beta, dout)\n\n_, cache = spatial_groupnorm_quick_forward(x, gamma, beta, G, gn_param)\ndx, dgamma, dbeta = spatial_groupnorm_backward(dout, cache)\n#You should expect errors of magnitudes between 1e-12~1e-07\nprint('dx error: ', rel_error(dx_num, dx))\nprint('dgamma error: ', rel_error(da_num, dgamma))\nprint('dbeta error: ', rel_error(db_num, dbeta))",
"dx error: 7.413109542981906e-08\ndgamma error: 9.468085754206675e-12\ndbeta error: 3.35440867127888e-12\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d00420c554adf6b85194a495ea114ccdcbbd6599 | 549,303 | ipynb | Jupyter Notebook | Experiment1_Main/Components/One25/one25.ipynb | ttrogers/frigo-chen-rogers | ddc8808f21a89259df83a161ee72faf2487623d4 | [
"MIT"
] | null | null | null | Experiment1_Main/Components/One25/one25.ipynb | ttrogers/frigo-chen-rogers | ddc8808f21a89259df83a161ee72faf2487623d4 | [
"MIT"
] | null | null | null | Experiment1_Main/Components/One25/one25.ipynb | ttrogers/frigo-chen-rogers | ddc8808f21a89259df83a161ee72faf2487623d4 | [
"MIT"
] | null | null | null | 79.643758 | 26,982 | 0.703996 | [
[
[
"# Pre-processing and analysis for one-source with distance 25",
"_____no_output_____"
],
[
"## Load or create R scripts",
"_____no_output_____"
]
],
[
[
"get.data <- dget(\"get_data.r\") #script to read data files\nget.pars <- dget(\"get_pars.r\") #script to extract relevant parameters from raw data\nget.mv.bound <- dget(\"get_mvbound.r\") #script to look at movement of boundary across learning\nplot.cirib <- dget(\"plot_cirib.r\") #script to plot confidence intervals as ribbon plot",
"_____no_output_____"
],
[
"zscore <- function(v){(v - mean(v, na.rm=T))/sqrt(var(v, na.rm=T))} #function to compute Z score",
"_____no_output_____"
]
],
[
[
"## Load data",
"_____no_output_____"
]
],
[
[
"fnames <- list.files(pattern = \"*.csv\") #create a vector of data file names, assuming all csv files are data\nnfiles <- length(fnames) #number of data files\nalldat <- list(get.data(fnames[1])) #initialize list containing all data with first subject\nfor(i1 in c(2:nfiles)) alldat[[i1]] <- get.data(fnames[i1]) #populate list with rest of data\n\nallpars <- get.pars(alldat) #extract parameters from grid test 1 and 2 from all data",
"[1] \"Processing sj: 1\"\n"
]
],
[
[
"NOTE that get pars will produce warnings whenever the subject data has a perfectly strict boundary--these can be safely ignored.",
"_____no_output_____"
]
],
[
[
"head(allpars)",
"_____no_output_____"
],
[
"dim(allpars)",
"_____no_output_____"
]
],
[
[
"### KEY",
"_____no_output_____"
],
[
"**PID**: Unique tag for each participant \n**cond**: Experiment condition \n**axlab**: What shape (spiky/smooth) got the \"Cooked\" label? For counterbalancing, not interesting \n**closebound**: What was the location of the closest source boundary? \n**cbside**: Factor indicating what side of the range midpoint has the close boundary\n**sno**: Subject number in condition \n**txint, slope, bound**: Intercept, slope, and estimated boundary from logistic regression on test 1 and test 2 data. NOTE that only the boundary estimate is used in analysis. \n**bshift**: Boundary shift direction and magnitude measured as test 2 boundary - test 1 boundary \n**alshift**: Placeholder for aligned boundary shift (see below), currently just a copy of bshift \n**Zalshift**: Zscored midshift, recalculated below",
"_____no_output_____"
],
[
"## Check data for outliers",
"_____no_output_____"
],
[
"Set Zscore threshold for outlier rejection",
"_____no_output_____"
]
],
[
[
"zthresh <- 2.5",
"_____no_output_____"
]
],
[
[
"First check t1bound and t2bound to see if there are any impossible values.",
"_____no_output_____"
]
],
[
[
"plot(allpars$t1bound, allpars$t2bound)",
"_____no_output_____"
]
],
[
[
"There is an impossible t2bound so let's remove it.",
"_____no_output_____"
]
],
[
[
"dim(allpars)\nsjex <- as.character(allpars$PID[allpars$t2bound < 0]) #Add impossible value to exclude list\nsjex <- unique(sjex) #remove any accidental repeats\nnoo <- allpars[is.na(match(allpars$PID, sjex)),] #Copy remaining subjects to noo object\ndim(noo)",
"_____no_output_____"
]
],
[
[
"Write \"no impossible\" (nimp) file for later agglomeration in mega-data",
"_____no_output_____"
]
],
[
[
"write.csv(noo, \"summary/one25_grids_nimp.csv\", row.names = F, quote=F)",
"_____no_output_____"
]
],
[
[
"Check to make sure \"aligned\" shift computation worked (should be an X pattern)",
"_____no_output_____"
]
],
[
[
"plot(noo$alshift, noo$bshift)",
"_____no_output_____"
]
],
[
[
"Check initial boundary for outliers",
"_____no_output_____"
]
],
[
[
"plot(zscore(noo$t1bound))\nabline(h=c(-zthresh,0,zthresh))",
"_____no_output_____"
]
],
[
[
"Add any outliers to the exclusion list and recompute no-outlier data structure",
"_____no_output_____"
]
],
[
[
"sjex <- c(sjex, as.character(allpars$PID[abs(zscore(allpars$t1bound)) > zthresh]))\nsjex <- unique(sjex) #remove accidental repeats\nnoo <- noo[is.na(match(noo$PID, sjex)),]\ndim(noo)",
"_____no_output_____"
]
],
[
[
"Now compute Zscore for aligned shift for all subjects and look for outliers",
"_____no_output_____"
]
],
[
[
"noo$Zalshift <- zscore(noo$alshift) #Compute Z scores for this aligned shift\nplot(noo$Zalshift); abline(h = c(-zthresh,0,zthresh)) #plot Zscores",
"_____no_output_____"
]
],
[
[
"Again add any outliers to exclusion list and remove from noo",
"_____no_output_____"
]
],
[
[
"sjex <- c(sjex, as.character(noo$PID[abs(noo$Zalshift) > zthresh]))\nsjex <- unique(sjex) #remove accidental repeats\nnoo <- noo[is.na(match(noo$PID, sjex)),]\ndim(noo)",
"_____no_output_____"
]
],
[
[
"## Data analysis",
"_____no_output_____"
],
[
"Does the initial (t1) boundary differ between the two groups? It shouldn't since they have the exact same experience to this point.",
"_____no_output_____"
]
],
[
[
"t.test(t1bound ~ closebound, data = noo)",
"_____no_output_____"
]
],
[
[
"Reassuringly, it doesn't. So what is the location of the initial boundary on average?",
"_____no_output_____"
]
],
[
[
"t.test(noo$t1bound) #NB t.test of a single vector is a good way to compute mean and CIs ",
"_____no_output_____"
]
],
[
[
"The mean boundary is shifted a bit positive relative to the midpoint between labeled examples\n\nNext, looking across all subjects, does the aligned boundary shift differ reliably from zero? Also, what are the confidence limits on the mean shift?",
"_____no_output_____"
]
],
[
[
"t.test(noo$alshift)",
"_____no_output_____"
]
],
[
[
"The boundary shifts reliably toward the close source. The mean amount of shift is 18, and the confidence interval spans 9-27.\n\nNext, where does the test 2 boundary lie for each group, and does this differ depending on where the source was?",
"_____no_output_____"
]
],
[
[
"t.test(t2bound ~ closebound, data = noo)",
"_____no_output_____"
]
],
[
[
"When the source was at 125, the boundary ends up at 134; when the source is at 175, the boundary ends up at 166.\n\nIs the boundary moving all the way to the source?",
"_____no_output_____"
]
],
[
[
"t.test(noo$t2bound[noo$closebound==125]) #compute confidence intervals for source at 125 subgroup",
"_____no_output_____"
],
[
"t.test(noo$t2bound[noo$closebound==175]) #compute confidence intervals for source at 175 subgroup",
"_____no_output_____"
]
],
[
[
"In both cases boundaries move toward the source. When the initial boundary is closer to the source (source at 175), the final boundary ends up at the source. When it is farther away (source at 125), the final boundary ends up a little short of the source.\n\nAnother way of looking at the movement is to compute, for each subject, how far the source was from the learner's initial boundary, and see if this predicts the amount of shift:",
"_____no_output_____"
]
],
[
[
"#Predict the boundary shift from the distance between initial bound and source\nm <- lm(bshift ~ t1dist, data = noo) #fit linear model predicting shift from distance\nsummary(m) #look at model parameters",
"_____no_output_____"
]
],
[
[
"Distance predicts shift significantly. The intercept is not reliably different from zero, so that, with zero distance, boundary does not shift. The slope of 0.776 suggests that the boundary shifts 78 percent of the way toward the close source. Let's visualize:",
"_____no_output_____"
]
],
[
[
"plot(noo$t1dist, noo$bshift) #plot distance of source against boundary shift\nabline(lm(bshift~t1dist, data = noo)$coefficients) #add least squares line\nabline(0,1, col = 2) #Add line with slope 1 and intercept 0 ",
"_____no_output_____"
]
],
[
[
"The black line shows the least-squares linear fit; the red line shows the expected slope if learner moved all the way toward the source. True slope is quite a bit shallower. If we compute confidence limits on slope we get:",
"_____no_output_____"
]
],
[
[
"confint(m, 't1dist', level = 0.95)",
"_____no_output_____"
]
],
[
[
"So the confidence limit extends v close to 1",
"_____no_output_____"
],
[
"### Export parameter data",
"_____no_output_____"
]
],
[
[
"write.csv(noo, paste(\"summary/onesrc25_noo_z\", zthresh*10, \".csv\", sep=\"\"), row.names=F, quote=F)",
"_____no_output_____"
]
],
[
[
"## Further analyses",
"_____no_output_____"
],
[
"### Movement of boundary over the course of learning",
"_____no_output_____"
]
],
[
[
"nsj <- length(alldat) #Number of subjects is length of alldat object\nmvbnd <- matrix(0, nsj, 301) #Initialize matrix of 0s to hold boundary-movement data, with 301 windows\nfor(i1 in c(1:nsj)) mvbnd[i1,] <- get.mv.bound(alldat, sj=i1) #Compute move data for each sj and store in matrix rows\n",
"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n\"glm.fit: algorithm did not converge\"Warning message:\n\"glm.fit: fitted probabilities numerically 0 or 1 occurred\"Warning message:\n"
]
],
[
[
"Again, ignore warnings here",
"_____no_output_____"
]
],
[
[
"tmp <- cbind(allpars[,1:6], mvbnd) #Add subject and condition data columns\nmvb.noo <- tmp[is.na(match(tmp$PID, sjex)),] #Remove excluded subjects",
"_____no_output_____"
],
[
"head(mvb.noo)",
"_____no_output_____"
],
[
"tmp <- mvb.noo[,7:307] #Copy movement data into temporary object\ntmp[abs(tmp) > 250] <- NA #Remove boundary estimates that are extreme (outside 50-250 range)\ntmp[tmp < 50] <- NA\nmvb.noo[,7:307] <- tmp #Put remaining data back in\n",
"_____no_output_____"
],
[
"plot.cirib(mvb.noo[mvb.noo$bounds==125,7:307], genplot=T)\nplot.cirib(mvb.noo[mvb.noo$bounds==175,7:307], genplot=F, color=4)\nabline(h=150, lty=2)\nabline(h=175, col=4)\nabline(h=125, col=2)\ntitle(\"Boundary shift over training\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d004241124de9de95e3a3f380317afae282cf966 | 126,007 | ipynb | Jupyter Notebook | Notes/KerasExercise.ipynb | GrayLand119/GLColabNotes | b42729491c20af0d9fb44c3de66f8b2756cccf03 | [
"MIT"
] | null | null | null | Notes/KerasExercise.ipynb | GrayLand119/GLColabNotes | b42729491c20af0d9fb44c3de66f8b2756cccf03 | [
"MIT"
] | null | null | null | Notes/KerasExercise.ipynb | GrayLand119/GLColabNotes | b42729491c20af0d9fb44c3de66f8b2756cccf03 | [
"MIT"
] | null | null | null | 152.735758 | 38,972 | 0.878308 | [
[
[
"# About\n\n此笔记包含了以下内容:\n\n* keras 的基本使用\n* 组合特征\n* 制作dataset\n* 模型的存取(2种方式)\n* 添加检查点\n",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom tensorflow.keras import layers\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport math",
"_____no_output_____"
],
[
"from tensorflow.keras.utils import plot_model\nimport os",
"_____no_output_____"
],
[
"# fea_x = [i for i in np.arange(0, math.pi * 2.0, 0.01)]\n# print(fea_x[:50])",
"_____no_output_____"
],
[
"x0 = np.random.randint(0, math.pi * 6.0 * 100.0, 5000) / 100.0\nx1 = np.random.randint(0, math.pi * 6.0 * 100.0, 5000) / 100.0\nx2 = np.random.randint(0, math.pi * 6.0 * 100.0, 1000) / 100.0 # Noisy\n\nfeaY0 = [np.random.randint(10 * math.sin(i), 20) for i in x0]\nfeaY1 = [np.random.randint(-20, 10 * math.sin(i)) for i in x1]\nfeaY2 = [np.random.randint(-10, 10) for i in x2]\n\nfea_x = np.concatenate([x0, x1, x2])\nfea_y = np.concatenate([feaY0, feaY1, feaY2])\n\nlabel0 = np.repeat(0, 5000)\nlabel1 = np.repeat(1, 5000)\nlabel2 = np.random.randint(0,2, 1000)\nlabel = np.concatenate([label0, label1, label2])\n",
"_____no_output_____"
],
[
"fea_1 = []\nfea_2 = []\nfea_3 = []\nfea_4 = []\nfea_5 = []\nfor i in range(len(label)):\n x = fea_x[i]\n y = fea_y[i]\n ex_1 = x * y\n ex_2 = x * x\n ex_3 = y * y\n ex_4 = math.sin(x)\n ex_5 = math.sin(y)\n fea_1.append(ex_1)\n fea_2.append(ex_2)\n fea_3.append(ex_3)\n fea_4.append(ex_4)\n fea_5.append(ex_5)\n ",
"_____no_output_____"
],
[
"fea = np.c_[fea_x, fea_y, fea_1, fea_2, fea_3, fea_4, fea_5]\ndataset = tf.data.Dataset.from_tensor_slices((fea, label))\ndataset = dataset.shuffle(10000)\ndataset = dataset.batch(500)\ndataset = dataset.repeat()\nds_iteror = dataset.make_one_shot_iterator().get_next()",
"WARNING: Logging before flag parsing goes to stderr.\nW0918 10:28:35.711431 4752016832 deprecation.py:323] From <ipython-input-5-ddde1f6f3511>:6: DatasetV1.make_one_shot_iterator (from tensorflow.python.data.ops.dataset_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse `for ... in dataset:` to iterate over a dataset. If using `tf.estimator`, return the `Dataset` object directly from your input function. As a last resort, you can use `tf.compat.v1.data.make_one_shot_iterator(dataset)`.\n"
],
[
"len(fea[0])",
"_____no_output_____"
],
[
"with tf.Session() as sess:\n def _pltfunc(sess):\n res = sess.run(ds_iteror)\n # print(res)\n lb = res[1]\n t_fea = res[0]\n for index in range(len(lb)):\n tfs = t_fea[index]\n if lb[index] > 0:\n plt.scatter(tfs[0], tfs[1], marker='o', c='orange')\n else:\n plt.scatter(tfs[0], tfs[1], marker='o', c='green')\n \n _pltfunc(sess)\n _pltfunc(sess)\n _pltfunc(sess)\n\n \nplt.show()",
"_____no_output_____"
],
[
"inputs = tf.keras.Input(shape=(7, ))\n\nx = layers.Dense(7, activation=tf.keras.activations.relu)(inputs)\nx1 = layers.Dense(7, activation='relu')(x)\n# x2 = layers.Dense(32, activation='relu')(x1)\n# x3 = layers.Dense(24, activation='relu')(x2)\n# x4 = layers.Dense(16, activation='relu')(x3)\n# x5 = layers.Dense(8, activation='relu')(x4)\npredictions = layers.Dense(2, activation='softmax')(x1)",
"W0918 10:28:44.855381 4752016832 deprecation.py:506] From /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages/tensorflow/python/ops/init_ops.py:1251: calling VarianceScaling.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\n"
],
[
"model = tf.keras.Model(inputs=inputs, outputs=predictions)\n\n# The compile step specifies the training configuration.\n# opt = tf.train.AdamOptimizer(learning_rate=0.0001)\nopt = tf.train.AdagradOptimizer(learning_rate=0.1)\n# opt = tf.train.RMSPropOptimizer(0.1)\nmodel.compile(optimizer=opt,\n loss=tf.keras.losses.sparse_categorical_crossentropy,\n metrics=['accuracy'])\n\nmodel.fit(dataset, epochs=10, steps_per_epoch=200)\n# model.fit(fea, label, epochs=10, batch_size=500, steps_per_epoch=300)",
"W0918 10:28:54.534353 4752016832 deprecation.py:506] From /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages/tensorflow/python/training/adagrad.py:76: calling Constant.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\n"
],
[
"model.fit(dataset, epochs=10, steps_per_epoch=200)",
"Epoch 1/10\n200/200 [==============================] - 0s 2ms/step - loss: 0.2353 - acc: 0.9100\nEpoch 2/10\n200/200 [==============================] - 0s 2ms/step - loss: 0.2321 - acc: 0.9133\nEpoch 3/10\n200/200 [==============================] - 0s 2ms/step - loss: 0.2279 - acc: 0.9157\nEpoch 4/10\n200/200 [==============================] - 0s 2ms/step - loss: 0.2253 - acc: 0.9177\nEpoch 5/10\n200/200 [==============================] - 0s 2ms/step - loss: 0.2238 - acc: 0.9190\nEpoch 6/10\n200/200 [==============================] - 0s 2ms/step - loss: 0.2241 - acc: 0.9200\nEpoch 7/10\n200/200 [==============================] - 0s 2ms/step - loss: 0.2228 - acc: 0.9214\nEpoch 8/10\n200/200 [==============================] - 0s 2ms/step - loss: 0.2220 - acc: 0.9222\nEpoch 9/10\n200/200 [==============================] - 0s 2ms/step - loss: 0.2212 - acc: 0.9219\nEpoch 10/10\n200/200 [==============================] - 0s 2ms/step - loss: 0.2204 - acc: 0.9233\n"
],
[
"result = model.predict([[[1, -10]]])\nprint(np.argmax(result[0]))\n\nresult = model.predict([[[1, 10]]])\nprint(np.argmax(result[0]))",
"1\n0\n"
],
[
"os.getcwd()",
"_____no_output_____"
],
[
"# 模型可视化\nplot_model(model, to_file=os.getcwd()+ '/model.png')",
"_____no_output_____"
],
[
"from IPython.display import SVG\nimport tensorflow.keras.utils as tfku\n\ntfku.plot_model(model)\n# SVG(model_to_dot(model).create(prog='dot', format='svg'))",
"_____no_output_____"
],
[
"for i in range(1000):\n randomX = np.random.randint(0, 10 * math.pi * 6.0) / 10.0\n randomY = 0\n if np.random.randint(2) > 0:\n randomY = np.random.randint(10 * math.sin(randomX), 20)\n else:\n randomY = np.random.randint(-20, 10 * math.sin(randomX))\n ex_1 = randomX * randomY\n ex_2 = randomX**2\n ex_3 = randomY**2\n ex_4 = math.sin(randomX)\n ex_5 = math.sin(randomY)\n \n color = ''\n result = model.predict([[[randomX, randomY, ex_1, ex_2, ex_3, ex_4, ex_5]]])\n pred_index = np.argmax(result[0])\n if pred_index > 0:\n color = 'orange'\n else:\n color = 'green'\n plt.scatter(randomX, randomY, marker='o', c=color)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Save Model",
"_____no_output_____"
]
],
[
[
"!pip install h5py pyyaml",
"Looking in indexes: https://pypi.tuna.tsinghua.edu.cn/simple\nRequirement already satisfied: h5py in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (2.9.0)\nRequirement already satisfied: pyyaml in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (5.1)\nRequirement already satisfied: six in /Users/languilin/Library/Python/3.6/lib/python/site-packages (from h5py) (1.12.0)\nRequirement already satisfied: numpy>=1.7 in /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages (from h5py) (1.16.2)\n\u001b[33mWARNING: You are using pip version 19.2.2, however version 19.2.3 is available.\nYou should consider upgrading via the 'pip install --upgrade pip' command.\u001b[0m\n"
],
[
"model_path = os.getcwd() + \"/mymodel.h5\"\nmodel_path",
"_____no_output_____"
]
],
[
[
"这里使用默认的优化器, 默认优化器不能直接保存, 读取模型时需要再次创建优化器并编译\n使用 keras 内置的优化器可以直接保存和读取, 比如: `tf.keras.optimizers.Adam()`",
"_____no_output_____"
]
],
[
[
"model.save(model_path)",
"W0919 14:36:37.931401 4752016832 hdf5_format.py:110] TensorFlow optimizers do not make it possible to access optimizer attributes or optimizer state after instantiation. As a result, we cannot save the optimizer as part of the model save file. You will have to compile your model again after loading it. Prefer using a Keras optimizer instead (see keras.io/optimizers).\n"
],
[
"new_model = tf.keras.models.load_model(model_path)\nopt = tf.train.AdagradOptimizer(learning_rate=0.1)\n# opt = tf.train.RMSPropOptimizer(0.1)\nnew_model.compile(optimizer=opt,\n loss=tf.keras.losses.sparse_categorical_crossentropy,\n metrics=['accuracy'])\n\nnew_model.summary()",
"W0919 14:43:45.045204 4752016832 deprecation.py:506] From /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages/tensorflow/python/ops/init_ops.py:97: calling GlorotUniform.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\nW0919 14:43:45.047919 4752016832 deprecation.py:506] From /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages/tensorflow/python/ops/init_ops.py:97: calling Zeros.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\nW0919 14:43:45.427887 4752016832 hdf5_format.py:221] No training configuration found in save file: the model was *not* compiled. Compile it manually.\n"
],
[
"loss, acc = new_model.evaluate(dataset, steps=200)\nprint(\"Restored model, accuracy: {:5.2f}%\".format(100*acc))\n",
"200/200 [==============================] - 1s 3ms/step - loss: 0.2201 - acc: 0.9251\nRestored model, accuracy: 92.51%\n"
],
[
"print(new_model.layers[1].get_weights())",
"[array([[-0.18513356, -0.5944598 , -0.31596622, -0.5336512 , 0.21966298,\n 0.4753986 , 0.41855925],\n [-0.51090246, -0.6128717 , 0.57087266, 0.90797025, -0.67894036,\n 0.12775406, -0.46071798],\n [-0.09616593, 0.18898904, 0.36915025, 0.22494109, -0.4048719 ,\n 0.08799099, -0.17466974],\n [ 0.47254115, -0.59851056, -0.64332885, -0.07513103, 0.27138317,\n -0.5164682 , 0.17601056],\n [ 0.4152183 , -0.5027053 , 0.18780918, 0.10085826, 0.04974491,\n -0.28167075, -0.01286947],\n [ 0.87135893, 0.6427965 , -0.28651774, -0.9457015 , 2.4377482 ,\n 0.35147208, -4.6972322 ],\n [-0.41829014, 0.22775614, 0.51005393, -0.78283525, 0.3037345 ,\n 0.23245245, -0.29947385]], dtype=float32), array([ 0.1523906 , 0.02099093, -0.01079311, 0.20105849, -0.00879347,\n -0.02339827, 0.32012293], dtype=float32)]\n"
],
[
"print(new_model.layers[3].get_weights())",
"[array([[ 0.06555444, -0.5212194 ],\n [-0.5949386 , -0.09970472],\n [-0.37951565, -0.21464492],\n [-0.13808419, 0.24510457],\n [ 0.36669165, -0.2663816 ],\n [ 0.45086718, -0.26410016],\n [-0.04899281, -0.6156222 ]], dtype=float32), array([-0.4162824, 0.4162828], dtype=float32)]\n"
]
],
[
[
"# 保存为 pb 文件",
"_____no_output_____"
]
],
[
[
"pb_model_path = os.getcwd() + '/pbmdoel'\npb_model_path",
"_____no_output_____"
],
[
"tf.contrib.saved_model.save_keras_model(new_model, pb_model_path)",
"W0919 14:54:26.258198 4752016832 deprecation.py:323] From /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages/tensorflow/python/saved_model/signature_def_utils_impl.py:253: build_tensor_info (from tensorflow.python.saved_model.utils_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis function will only be available through the v1 compatibility library as tf.compat.v1.saved_model.utils.build_tensor_info or tf.compat.v1.saved_model.build_tensor_info.\nW0919 14:54:26.259163 4752016832 export_utils.py:182] Export includes no default signature!\nW0919 14:54:26.461674 4752016832 export_utils.py:182] Export includes no default signature!\n"
],
[
"!ls {pb_model_path}",
"\u001b[34massets\u001b[m\u001b[m saved_model.pb \u001b[34mvariables\u001b[m\u001b[m\r\n"
]
],
[
[
"# 读取 pb 文件",
"_____no_output_____"
]
],
[
[
"model2 = tf.contrib.saved_model.load_keras_model(pb_model_path)\nmodel2.summary()",
"Model: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 7)] 0 \n_________________________________________________________________\ndense (Dense) (None, 7) 56 \n_________________________________________________________________\ndense_1 (Dense) (None, 7) 56 \n_________________________________________________________________\ndense_2 (Dense) (None, 2) 16 \n=================================================================\nTotal params: 128\nTrainable params: 128\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# 使用前要先编译\nmodel2.compile(optimizer=opt,\n loss=tf.keras.losses.sparse_categorical_crossentropy,\n metrics=['accuracy'])\n\nloss, acc = model2.evaluate(dataset, steps=200)\nprint(\"Restored model, accuracy: {:5.2f}%\".format(100*acc))",
"200/200 [==============================] - 0s 2ms/step - loss: 0.2203 - acc: 0.9250\nRestored model, accuracy: 92.50%\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0042eab5854b447de51a429a272d3a09f8991fe | 10,515 | ipynb | Jupyter Notebook | 2016/loris/day_1.ipynb | bbglab/adventofcode | 65b6d8331d10f229b59232882d60024b08d69294 | [
"MIT"
] | null | null | null | 2016/loris/day_1.ipynb | bbglab/adventofcode | 65b6d8331d10f229b59232882d60024b08d69294 | [
"MIT"
] | null | null | null | 2016/loris/day_1.ipynb | bbglab/adventofcode | 65b6d8331d10f229b59232882d60024b08d69294 | [
"MIT"
] | 3 | 2016-12-02T09:20:42.000Z | 2021-12-01T13:31:07.000Z | 36.510417 | 277 | 0.463338 | [
[
[
"# Advent of Code 2016",
"_____no_output_____"
]
],
[
[
"--- Day 1: No Time for a Taxicab ---\n\nSanta's sleigh uses a very high-precision clock to guide its movements, and the clock's oscillator is regulated by stars. Unfortunately, the stars have been stolen... by the Easter Bunny. To save Christmas, Santa needs you to retrieve all fifty stars by December 25th.\n\nCollect stars by solving puzzles. Two puzzles will be made available on each day in the advent calendar; the second puzzle is unlocked when you complete the first. Each puzzle grants one star. Good luck!\n\nYou're airdropped near Easter Bunny Headquarters in a city somewhere. \"Near\", unfortunately, is as close as you can get - the instructions on the Easter Bunny Recruiting Document the Elves intercepted start here, and nobody had time to work them out further.\n\nThe Document indicates that you should start at the given coordinates (where you just landed) and face North. Then, follow the provided sequence: either turn left (L) or right (R) 90 degrees, then walk forward the given number of blocks, ending at a new intersection.\n\nThere's no time to follow such ridiculous instructions on foot, though, so you take a moment and work out the destination. Given that you can only walk on the street grid of the city, how far is the shortest path to the destination?\n\nFor example:\n\nFollowing R2, L3 leaves you 2 blocks East and 3 blocks North, or 5 blocks away.\nR2, R2, R2 leaves you 2 blocks due South of your starting position, which is 2 blocks away.\nR5, L5, R5, R3 leaves you 12 blocks away.\n\nHow many blocks away is Easter Bunny HQ?",
"_____no_output_____"
]
],
[
[
"data = open('data/day_1-1.txt', 'r').readline().strip().split(', ')",
"_____no_output_____"
],
[
"class TaxiCab:\n \n def __init__(self, data):\n self.data = data\n self.double_visit = []\n self.position = {'x': 0, 'y': 0}\n self.direction = {'x': 0, 'y': 1}\n self.grid = {i: {j: 0 for j in range(-500, 501)} for i in range(-500, 501)}\n \n def run(self):\n for instruction in self.data:\n toward = instruction[0]\n length = int(instruction[1:])\n self.move(toward, length)\n \n def move(self, toward, length):\n if toward == 'R':\n if self.direction['x'] == 0:\n # from UP\n if self.direction['y'] == 1:\n self.position['x'] += length\n self.direction['x'] = 1\n for i in range(self.position['x'] - length, self.position['x']):\n self.grid[self.position['y']][i] += 1\n if self.grid[self.position['y']][i] > 1:\n self.double_visit.append((i, self.position['y']))\n # from DOWN\n else:\n self.position['x'] -= length\n self.direction['x'] = -1\n for i in range(self.position['x'] + length, self.position['x'], -1):\n self.grid[self.position['y']][i] += 1\n if self.grid[self.position['y']][i] > 1:\n self.double_visit.append((i, self.position['y']))\n self.direction['y'] = 0\n else: \n # FROM RIGHT\n if self.direction['x'] == 1:\n self.position['y'] -= length\n self.direction['y'] = -1\n for i in range(self.position['y'] + length, self.position['y'], -1):\n self.grid[i][self.position['x']] += 1\n if self.grid[i][self.position['x']] > 1:\n self.double_visit.append((self.position['x'], i))\n # FROM LEFT\n else:\n self.position['y'] += length\n self.direction['y'] = 1\n for i in range(self.position['y'] - length, self.position['y']):\n self.grid[i][self.position['x']] += 1\n if self.grid[i][self.position['x']] > 1:\n self.double_visit.append((self.position['x'], i))\n self.direction['x'] = 0\n else:\n if self.direction['x'] == 0:\n # from UP\n if self.direction['y'] == 1:\n self.position['x'] -= length\n self.direction['x'] = -1\n for i in range(self.position['x'] + length, self.position['x'], -1):\n self.grid[self.position['y']][i] += 1\n if self.grid[self.position['y']][i] > 1:\n self.double_visit.append((i, self.position['y']))\n # from DOWN\n else:\n self.position['x'] += length\n self.direction['x'] = 1\n for i in range(self.position['x'] - length, self.position['x']):\n self.grid[self.position['y']][i] += 1\n if self.grid[self.position['y']][i] > 1:\n self.double_visit.append((i, self.position['y']))\n self.direction['y'] = 0\n else: \n # FROM RIGHT\n if self.direction['x'] == 1:\n self.position['y'] += length\n self.direction['y'] = 1\n for i in range(self.position['y'] - length, self.position['y']):\n self.grid[i][self.position['x']] += 1\n if self.grid[i][self.position['x']] > 1:\n self.double_visit.append((self.position['x'], i))\n # FROM LEFT\n else:\n self.position['y'] -= length\n self.direction['y'] = -1\n for i in range(self.position['y'] + length, self.position['y'], -1):\n self.grid[i][self.position['x']] += 1\n if self.grid[i][self.position['x']] > 1:\n self.double_visit.append((self.position['x'], i))\n self.direction['x'] = 0\n \n def get_distance(self):\n return sum([abs(i) for i in self.position.values()])\n \n def get_distance_first_double_visit(self):\n return sum(self.double_visit[0]) if len(self.double_visit) > 0 else 0",
"_____no_output_____"
],
[
"# Test \ndef test(data, result):\n tc = TaxiCab(data)\n tc.run()\n assert tc.get_distance() == result",
"_____no_output_____"
],
[
"test(data=['R2', 'L3'], result=5)\ntest(data=['R2', 'R2', 'R2'], result=2)\ntest(data=['R5', 'L5', 'R5', 'R3'], result=12)",
"_____no_output_____"
],
[
"tc = TaxiCab(data)\ntc.run()\ntc.get_distance()",
"_____no_output_____"
]
],
[
[
"--- Part Two ---\n\nThen, you notice the instructions continue on the back of the Recruiting Document. Easter Bunny HQ is actually at the first location you visit twice.\n\nFor example, if your instructions are R8, R4, R4, R8, the first location you visit twice is 4 blocks away, due East.\n\nHow many blocks away is the first location you visit twice?",
"_____no_output_____"
]
],
[
[
"# Test \ndef test(data, result):\n tc = TaxiCab(data)\n tc.run()\n assert tc.get_distance_first_double_visit() == result",
"_____no_output_____"
],
[
"test(data=['R8', 'R4', 'R4', 'R8'], result=4)",
"_____no_output_____"
],
[
"tc.get_distance_first_double_visit()",
"_____no_output_____"
]
]
] | [
"markdown",
"raw",
"code",
"raw",
"code"
] | [
[
"markdown"
],
[
"raw"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"raw"
],
[
"code",
"code",
"code"
]
] |
d004339a3b87926b5dad0ec695083c16c43f98ab | 4,990 | ipynb | Jupyter Notebook | Distance.ipynb | iscanegemen/Data-Science-with-Foursquare-API- | 8523509275a1abd6a2598746f86b2dfb33750b5a | [
"MIT"
] | 1 | 2020-11-26T17:33:28.000Z | 2020-11-26T17:33:28.000Z | Distance.ipynb | iscanegemen/Data-Science-with-Foursquare-API | 8523509275a1abd6a2598746f86b2dfb33750b5a | [
"MIT"
] | null | null | null | Distance.ipynb | iscanegemen/Data-Science-with-Foursquare-API | 8523509275a1abd6a2598746f86b2dfb33750b5a | [
"MIT"
] | null | null | null | 39.92 | 1,137 | 0.593988 | [
[
[
"import pandas as pd\nimport math\n\n\n\ndf=pd.read_csv(r\"C:\\Users\\MONSTER\\Desktop\\newyorkcoffeewithdetails.csv\",error_bad_lines=False)\n\ndistance_dict = {}\n\nlat_input=float(input(\"Latitude : \")) # User's latitude and longitude\nlon_input=float(input(\"longitude : \"))\n\n\nfor i in range(len(df)): # I take latitutude and longitude for every buisness\n \n # our latitude and longitude of surroindings\n lat = math.radians(df[\"location.lat\"].iloc[i])\n lon = math.radians(df[\"location.lng\"].iloc[i])\n \n R = 6373.0 # Diameter of earth\n \n distance_lon = lon - lon_input\n distance_lat = lat - lat_input\n \n #Haversine Formula for finding distance between two geographical locations in earth\n \n a = math.sin(distance_lat/ 2)**2 + math.cos(lat_input) * math.cos(lat) * math.sin(distance_lon / 2)**2\n c = 2 * math.atan2(math.sqrt(a), math.sqrt(1 - a))\n \n distance = R * c\n \n distance_dict[i]=distance\n \nsorted_values = sorted(distance_dict.values()) # Sort the values\nsorted_distance_dict = {}\n\n# I sort my dictionary in terms of values from small to big\nfor i in sorted_values:\n for k in distance_dict.keys():\n if distance_dict[k] == i:\n sorted_distance_dict[k] = i\n break\n\nprint(sorted_distance_dict)\n\nlocation_stats = pd.DataFrame(columns=[\"NAME\",\"LATITUDE\",\"LONGITUDE\",\"DISTANCE\"])\n\nindex_number_list = []\ndistances_list = []\n\nfor i in range(1,6):\n index_number_list.append(list(sorted_distance_dict.keys())[i])\n distances_list.append(sorted_distance_dict[list(sorted_distance_dict.keys())[i]])\n\n\nfor j in range(len(distances_list)):\n location_stats=location_stats.append({\"LATITUDE\":df[\"location.lat\"].iloc[index_number_list[j]],\"LONGITUDE\":df[\"location.lng\"].iloc[index_number_list[j]],\"NAME\": df[\"name\"].iloc[index_number_list[j]],\"DISTANCE\": distances_list[j]},ignore_index =True)\n\n\nprint(location_stats.head())\n\n ",
"Latitude : 12\nlongitude : 24\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
d0044a2c29cd84a8d9336751d88f257bba7451b1 | 14,085 | ipynb | Jupyter Notebook | notebooks/beginner/notebooks/strings.ipynb | jordantcarlisle/learn-python3 | 53964f7d67d64af10233f91403e04bb4d9b1a566 | [
"MIT"
] | null | null | null | notebooks/beginner/notebooks/strings.ipynb | jordantcarlisle/learn-python3 | 53964f7d67d64af10233f91403e04bb4d9b1a566 | [
"MIT"
] | null | null | null | notebooks/beginner/notebooks/strings.ipynb | jordantcarlisle/learn-python3 | 53964f7d67d64af10233f91403e04bb4d9b1a566 | [
"MIT"
] | null | null | null | 20.472384 | 224 | 0.496557 | [
[
[
"# [Strings](https://docs.python.org/3/library/stdtypes.html#text-sequence-type-str)",
"_____no_output_____"
]
],
[
[
"my_string = 'Python is my favorite programming language!'",
"_____no_output_____"
],
[
"my_string",
"_____no_output_____"
],
[
"type(my_string)",
"_____no_output_____"
],
[
"len(my_string)",
"_____no_output_____"
]
],
[
[
"## Respecting [PEP8](https://www.python.org/dev/peps/pep-0008/#maximum-line-length) with long strings",
"_____no_output_____"
]
],
[
[
"long_story = ('Lorem ipsum dolor sit amet, consectetur adipiscing elit.' \n 'Pellentesque eget tincidunt felis. Ut ac vestibulum est.' \n 'In sed ipsum sit amet sapien scelerisque bibendum. Sed ' \n 'sagittis purus eu diam fermentum pellentesque.')\nlong_story",
"_____no_output_____"
]
],
[
[
"## `str.replace()`",
"_____no_output_____"
],
[
"If you don't know how it works, you can always check the `help`:",
"_____no_output_____"
]
],
[
[
"help(str.replace)",
"Help on method_descriptor:\n\nreplace(self, old, new, count=-1, /)\n Return a copy with all occurrences of substring old replaced by new.\n \n count\n Maximum number of occurrences to replace.\n -1 (the default value) means replace all occurrences.\n \n If the optional argument count is given, only the first count occurrences are\n replaced.\n\n"
]
],
[
[
"This will not modify `my_string` because replace is not done in-place.",
"_____no_output_____"
]
],
[
[
"my_string.replace('a', '?')\nprint(my_string)",
"Python is my favorite programming language!\n"
]
],
[
[
"You have to store the return value of `replace` instead.",
"_____no_output_____"
]
],
[
[
"my_modified_string = my_string.replace('is', 'will be')\nprint(my_modified_string)",
"Python will be my favorite programming language!\n"
]
],
[
[
"## `str.format()`",
"_____no_output_____"
]
],
[
[
"secret = '{} is cool'.format('Python')\nprint(secret)",
"Python is cool\n"
],
[
"print('My name is {} {}, you can call me {}.'.format('John', 'Doe', 'John'))\n# is the same as:\nprint('My name is {first} {family}, you can call me {first}.'.format(first='John', family='Doe'))",
"My name is John Doe, you can call me John.\nMy name is John Doe, you can call me John.\n"
]
],
[
[
"## `str.join()`",
"_____no_output_____"
]
],
[
[
"help(str.join)",
"Help on method_descriptor:\n\njoin(self, iterable, /)\n Concatenate any number of strings.\n \n The string whose method is called is inserted in between each given string.\n The result is returned as a new string.\n \n Example: '.'.join(['ab', 'pq', 'rs']) -> 'ab.pq.rs'\n\n"
],
[
"pandas = 'pandas'\nnumpy = 'numpy'\nrequests = 'requests'\ncool_python_libs = ', '.join([pandas, numpy, requests])",
"_____no_output_____"
],
[
"print('Some cool python libraries: {}'.format(cool_python_libs))",
"Some cool python libraries: pandas, numpy, requests\n"
]
],
[
[
"Alternatives (not as [Pythonic](http://docs.python-guide.org/en/latest/writing/style/#idioms) and [slower](https://waymoot.org/home/python_string/)):",
"_____no_output_____"
]
],
[
[
"cool_python_libs = pandas + ', ' + numpy + ', ' + requests\nprint('Some cool python libraries: {}'.format(cool_python_libs))\n\ncool_python_libs = pandas\ncool_python_libs += ', ' + numpy\ncool_python_libs += ', ' + requests\nprint('Some cool python libraries: {}'.format(cool_python_libs))",
"Some cool python libraries: pandas, numpy, requests\nSome cool python libraries: pandas, numpy, requests\n"
]
],
[
[
"## `str.upper(), str.lower(), str.title()`",
"_____no_output_____"
]
],
[
[
"mixed_case = 'PyTHoN hackER'",
"_____no_output_____"
],
[
"mixed_case.upper()",
"_____no_output_____"
],
[
"mixed_case.lower()",
"_____no_output_____"
],
[
"mixed_case.title()",
"_____no_output_____"
]
],
[
[
"## `str.strip()`",
"_____no_output_____"
]
],
[
[
"help(str.strip)",
"Help on method_descriptor:\n\nstrip(self, chars=None, /)\n Return a copy of the string with leading and trailing whitespace removed.\n \n If chars is given and not None, remove characters in chars instead.\n\n"
],
[
"ugly_formatted = ' \\n \\t Some story to tell '\nstripped = ugly_formatted.strip()\n\nprint('ugly: {}'.format(ugly_formatted))\nprint('stripped: {}'.format(ugly_formatted.strip()))",
"ugly: \n \t Some story to tell \nstripped: Some story to tell\n"
]
],
[
[
"## `str.split()`",
"_____no_output_____"
]
],
[
[
"help(str.split)",
"Help on method_descriptor:\n\nsplit(self, /, sep=None, maxsplit=-1)\n Return a list of the words in the string, using sep as the delimiter string.\n \n sep\n The delimiter according which to split the string.\n None (the default value) means split according to any whitespace,\n and discard empty strings from the result.\n maxsplit\n Maximum number of splits to do.\n -1 (the default value) means no limit.\n\n"
],
[
"sentence = 'three different words'\nwords = sentence.split()\nprint(words)",
"['three', 'different', 'words']\n"
],
[
"type(words)",
"_____no_output_____"
],
[
"secret_binary_data = '01001,101101,11100000'\nbinaries = secret_binary_data.split(',')\nprint(binaries)",
"['01001', '101101', '11100000']\n"
]
],
[
[
"## Calling multiple methods in a row",
"_____no_output_____"
]
],
[
[
"ugly_mixed_case = ' ThIS LooKs BAd '\npretty = ugly_mixed_case.strip().lower().replace('bad', 'good')\nprint(pretty)",
"this looks good\n"
]
],
[
[
"Note that execution order is from left to right. Thus, this won't work:",
"_____no_output_____"
]
],
[
[
"pretty = ugly_mixed_case.replace('bad', 'good').strip().lower()\nprint(pretty)",
"this looks bad\n"
]
],
[
[
"## [Escape characters](http://python-reference.readthedocs.io/en/latest/docs/str/escapes.html#escape-characters)",
"_____no_output_____"
]
],
[
[
"two_lines = 'First line\\nSecond line'\nprint(two_lines)",
"First line\nSecond line\n"
],
[
"indented = '\\tThis will be indented'\nprint(indented)",
"\tThis will be indented\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |